What is Chromosomal Aberration?
Any type of change in the structure and number of chromosomes due to certain irregularities during cell division is known as chromosomal aberration.
Broadly such aberrations are divided into two major groups.
- Structural aberration
- Numerical aberration
Again, structural aberrations are of 4 types. Such as:
- Deletion or deficiency
- Duplication
- Inversion
- Translocation
Numerical chromosomal aberrations are of 2 types. Such as:
- Aneuploidy
- Euploidy
Structural chromosomal aberration
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
Any type of change in the structure of chromosomes due to certain irregularities during cell division is known as structural chromosomal aberration.
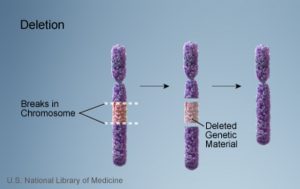
1a. Deletion
- Loss of segement or segments of a chromosome.
- Example: Wolf-Hirschhorn syndrome, which is caused by partial deletion of the short arm of chromosome 4; and Jacobsen syndrome, also called the terminal 11q deletion disorder.
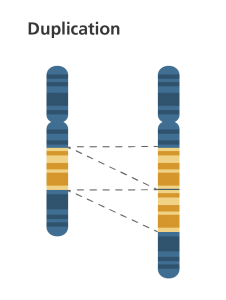
1b. Duplication
- A portion of the chromosome is duplicated, resulting in extra genetic material.
- The extra piece may get attatched to the same homologous chromosome or transposed to one of the nonhomologous members of the genome.
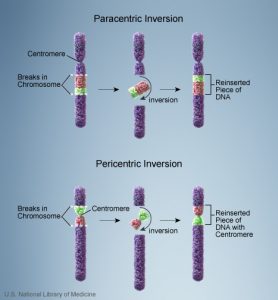
1c. Inversion
- A change in the linear sequence of the genes in a chromosome that results in the reverse order of the genes in a chromosome segment is called inversion. Or, simply put as, a portion of the chromosome has broken off, turned upside down, and reattached, therefore the genetic material is inverted.
- Inversions are widespread in plants, insects and mammals, and are found in nature or induced by radiation and chemical mutagens.
- Here, chromosomal breakage is followed by healing process.
- Inversions are responsible for speciations (Stebbins, 1950).
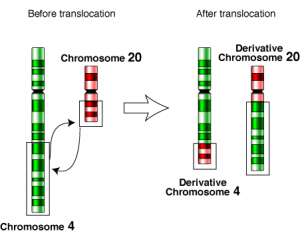
1d. Translocation
- Also called segmental chromosomal interchanges.
- It is the result of the reciprocal exchange of terminal segments of nonhomologous chromosomes.
Difference between crossing over and translocation
| Crossing Over | Translocation |
| Between homologous chromosomes. | Between non-homologous chromosomes. |
| Could be terminal or intercalary. | Always terminal. |
Numerical chromosomal aberration
Any type of change in the number of chromosomes due to certain irregularities during cell division is known as numerical chromosomal aberration.
An individual carrying chromosome numbers other than true monoploid or diploid numbers is called heteroploid. Heteroploidy is divided into aneuploify and euploidy.
2a. Aneuploidy: Change of chromosome number but not in a set. Here the chromosome number is some number other than the exact multiple of the basic chromosome set.
2b. Euploidy: Change of chromosome number in a set. So a euploid organism carries an exact multiple of the basic chromosome number.
Good to know:
- In polyploids, x is the basic (monoploid) chromosome number. n is the gametic chromosome number of chromosomes, and 2n is the zygotic or somatic chromosome number.
For example, the genomic formula of Triticum aestivum is 2n=6x=42 and Hordeum vulgare is 2n=2x=14. In both cases, the basic chromsome number x is seven.
- The basic set of chromsome in a diploid is called a genome.
Aneuploidy
Aneuploidy is again divided into two categories. Such as:
Hyper aneuploidy: In this type of aneuploidy, the chromosome number increases but not in a set. Such as:
- Trisomy: 2n+1
- Tetrasomy: 2n+2
- Double trisomy: 2n+1+1
Hypo aneuploidy: In this type of aneuploidy, the chromosome number decreases but not in a set. Such as:
- Monosomy: 2n-1
- Nullisomy: 2n-2
- Double monosomy: 2n-1-1
Euploidy
Euploidy is divided into two categories. Such as: Polyploid and Haploid
Polyploid: This aberration causes increase in the number of chromosomal set. It can be represented (Not types) as quantitative, qualitative and quantiqualitative.
Polyploidy is mainly divided into 4 categories such as:
- Autopolyploidy,
- Allopolyploidy,
- Autoallopolyploidy and
- Segmental allopolyploidy which are discussed under qualitative section.
Quantitative:
- Triploid: 3X
- Tetraploid: 4X
- Pentaploid: 5X and so on.
Qualitative:
- Autopolyploid: AAAA
- Allopolyploid: ABCD
- Autoallopolyploid: AAAABB (Presence of different types of genomes in which one genome remains as autopolyploid).
- Segmental allopolyploid: AA1B1B2C2C
Quantiqualitative:
- Allotriploid: ABC
- Autotetraploid: BBBB
- Segmental allohexaploid: B1B1D1D2C2C
- Autopolyploidy
- Here, all the genomes are alike.
- Eg. AAAA.
- One basic genome is multiplied (x, monoploid; 2x, diploid; 3x, triploid, 4x, tetraploid, 5x, pentaploid and so on).
2. Allopolyploidy
- Two or more genomes derived from different genomically unlike, distinct species are present.
- Eg. ABCD
3. Segmental allopolyploidy
- Carry genomes intermediate in degree of similarity and generally exhibit preferential pairing.
- For example: a segmental allotetraploid with genomes B1B1B2B2 usually forms bivalents and occassionally quadrivalents.
4. Autoallopolyploidy
- Confined to hexaploidy and higher levels of polyploidy (Stebbins, 1950).
- Eg. AAAABB
Significance of polyploidy
Polyploidy has played a major role in the speciation of higher plants. The species in which the basic chromsome number is x=10, or higher, evolved by polyploidization.
Haploid: Here chromosome decreases in set. It indicates that aberration where gametic cell is present in somatic body.
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka