DNA damage is defined as an alternative to DNA structures that can cause cellular injury and reduces the organisms efficiency or reproductive fitness. DNA damage may occur by free radical, physical agents like UV rays, X rays and chemical agents like aflatoxine, alkylating agents, hydraxines. Further, DNA damage is intertwined in mutagenesis and the development of cancer.
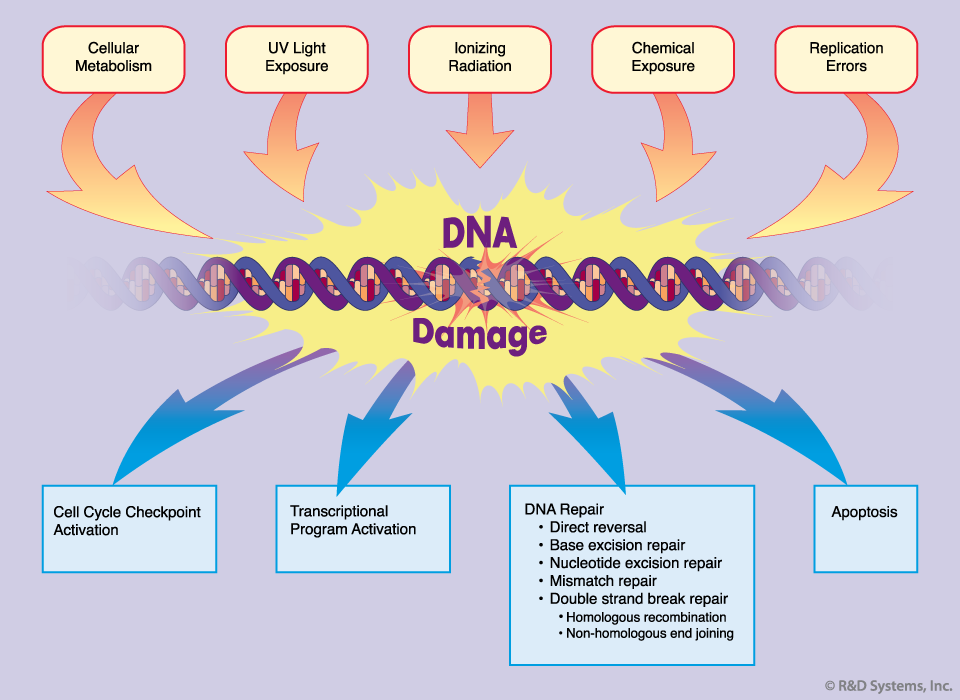
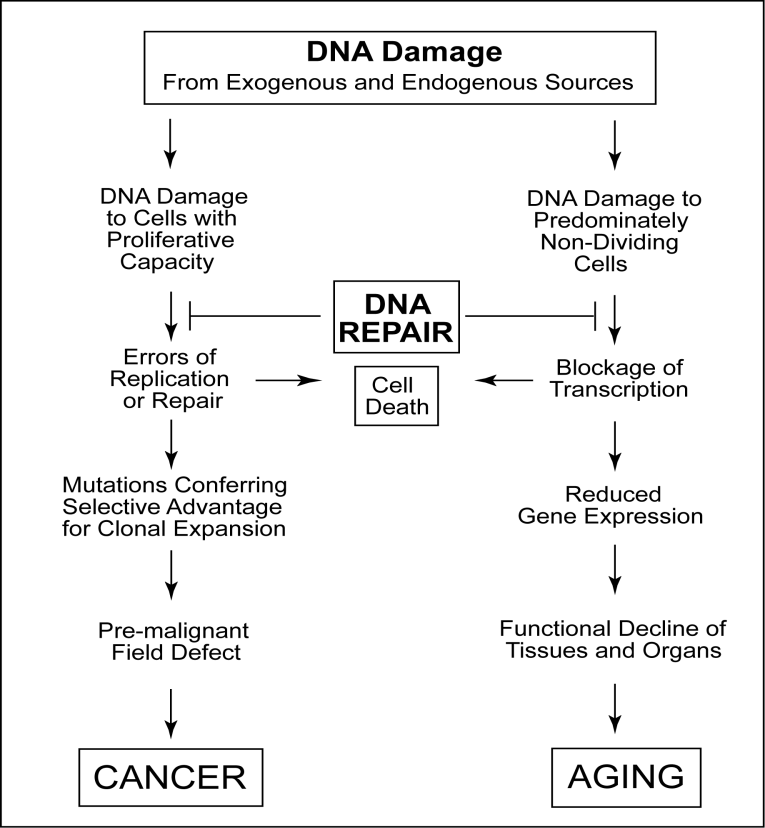
Types of DNA damage
DNA damage can be divided into two main classes based on its origin.
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
- Endogenous: Endogenous DNA damage include hydrolysis, oxidation, alkylation and mismatch of DNA bases.
- Exogenous: Exogenous DNA damage include ionizing radiation(IR), ultraviolet(UV) radiation, and various chemical agents.
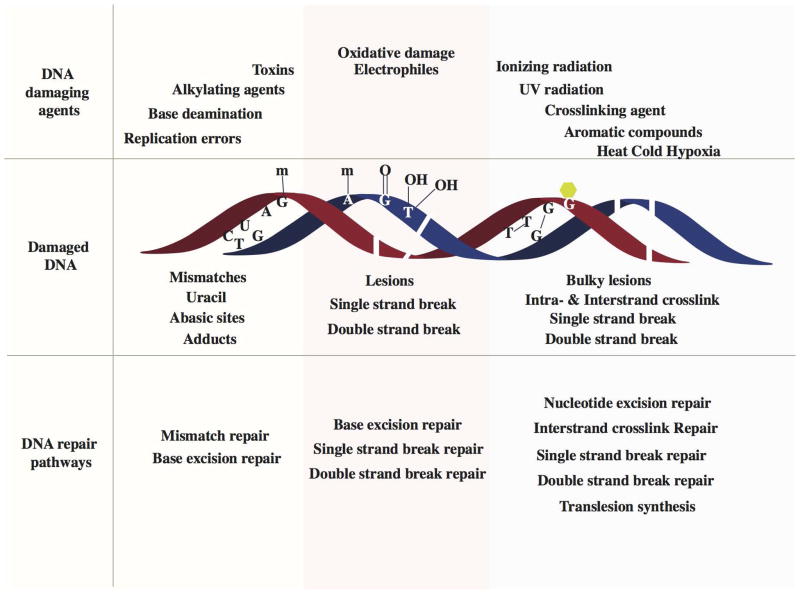
DNA repair is a collection of processes that detect the loss of DNA molecules and encode a cell’s genome. In human cells, both normal metabolic activity such as radiation and environmental factors can cause DNA damage. Molecular lesions can cause structural damage to DNA molecules. And it’s alter or remove the ability of cells to replicate genes that encode infected DNA. Other lesions incline possibly harmful mutations in the cell genome that affect the survival of its daughter cells after mitosis.
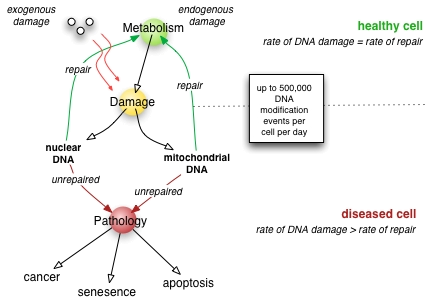
DNA repair can be divided into a process that identify and correct damage in DNA molecules. There are three kinds of DNA repair mechanisms. These are:
1. Direct reversal: Direct reversal repair is specific to the damage.
Photoreactivation
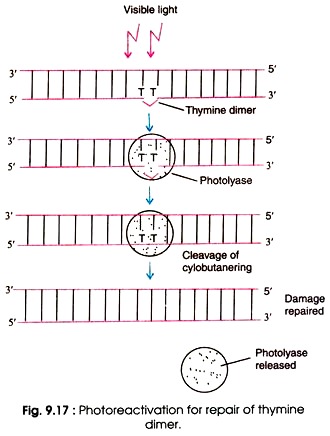
Photoreactivation is likely the simplest DNA repair mechanism which is present in prokaryotes, archaea and in many eukaryotes. It is lightly induced enzymatic fission of a thymine dimer to produce two thymine monomers. It is accomplished by photolyase enzyme that acts on dimers contained in single and double stranded DNA. Photolyase enzymes are found in fungi, plants, bacteria and vertebrate but are not found in mammals.
Hermann J. Muller demonstrates that X rays can induce mutations and can kill the cells. But later, other types of radiations including UV rays also effect cell function and mutations levels. In 1940s, Albert Kelner discovered that the UV reduced cellular damage that can be recovered by treating the cell with the visible light. And he called this process is ”Photoreactivation”
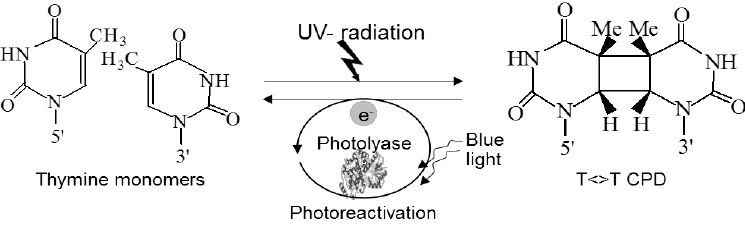
Mechanism of photoreactivation
- Firstly the photolase enzyme recognizes the thymine dimer and bind this thymine dymer.
- The enzyme absorbs visible light (300-500)nm
- After absorbing light, the FADH, cofactor get exited.
- The excited FADH then transfers the electron to the thymine dimer.
- As a result, charged redical pair is produced.
- Ring splits by cycloreversion.
- Connecting bonds break.
- Then Electron returns to the FADH.
- Phytolase leaves the DNA.
- Alkylation
Direct reversal of alkylation events, a DNA methyltransferase or DNA glycosylase detects and dispels the alkyl group.
2. Single strand repair
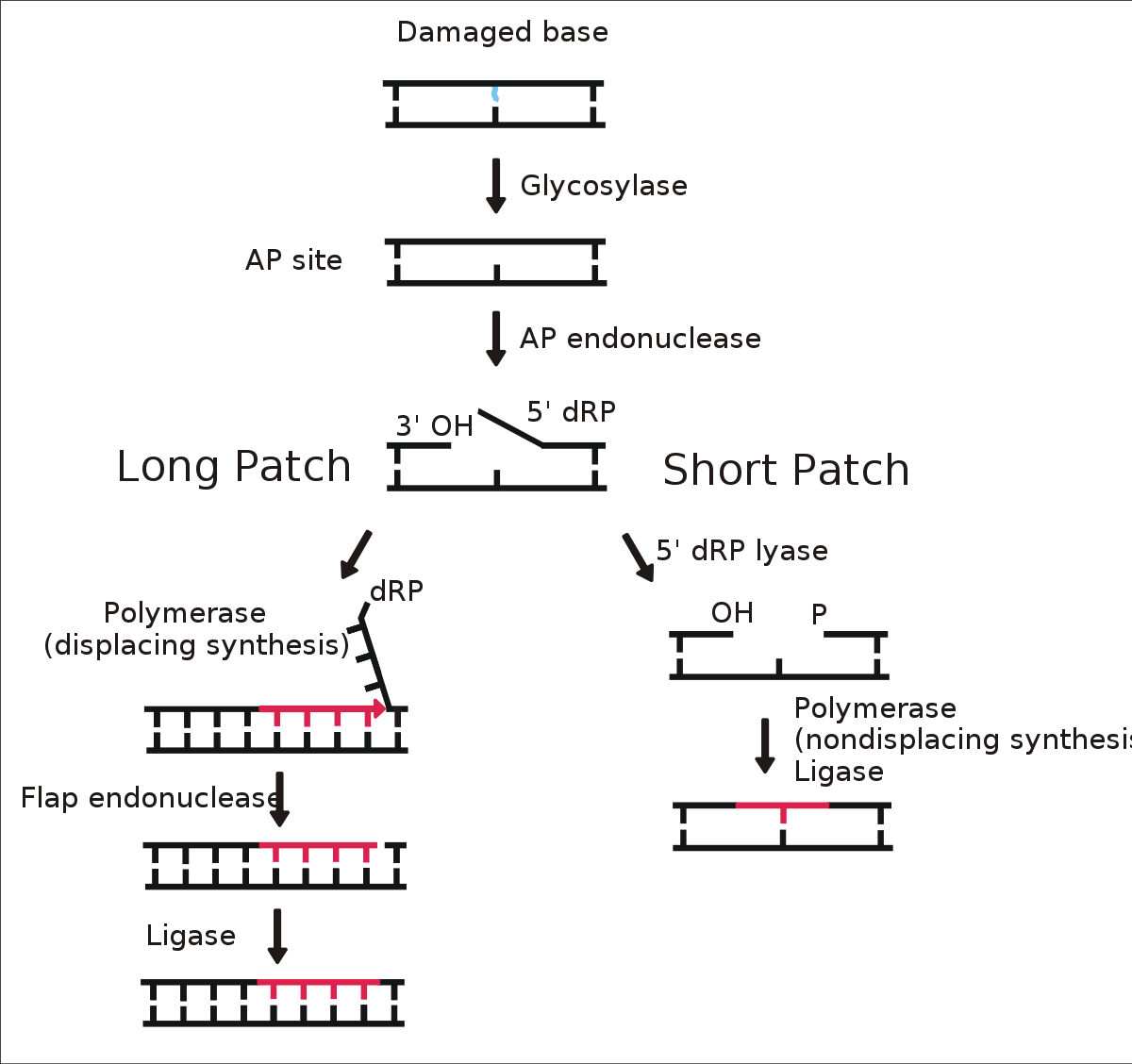
Base excision repair
Base-excision repair comprises the recognition and removal of a single damaged base. This process requires a family of enzymes called glycosylases. The enzymes take aside the damaged base forming an AP site which is repaired by AP endonuclease before the nucleotide gap in the DNA strand is filled by DNA polymerase.
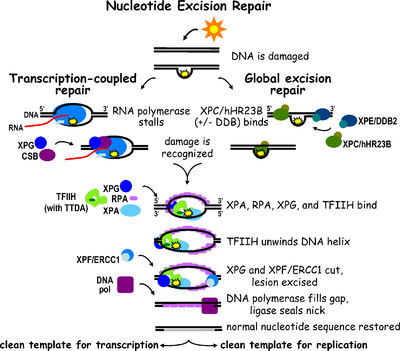
Nucleotide excision repair
Nucleotide excision repair is a comprehensive mechanism for repairing damage to DNA and detect multiple damaged bases. This process is used to repair the formation of pyrimidine dimers from UV light in humans. The process involves the recognition of damage that is then cleaved between the two sides by endonucleases before being resynthesized by DNA polymerase.
Mismatch repair
The third excision mechanism is called mismatch repair and it occurs when mismatched bases are attached to the DNA strand during replication and are not dispelled by proofreading DNA polymerase. In repair the discrepancy, the missed errors are then corrected by enzymes that detect and excise the mismatched base to restore the original sequence.
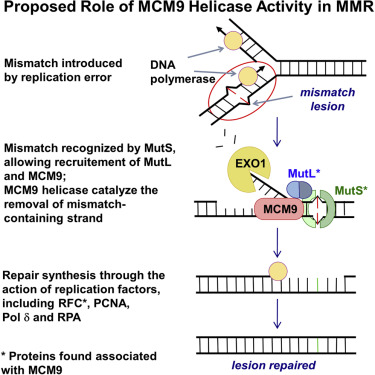
3.Double strand repair
DNA double-strand breaks are repaired through two main process. These are nonhomologous end joining and homologous recombination. Its also can be repaired by means of single-strand annealing between adjacent repeated DNA sequences, which involves deletion of the intervening DNA.
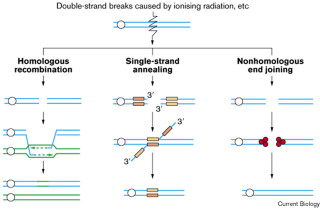
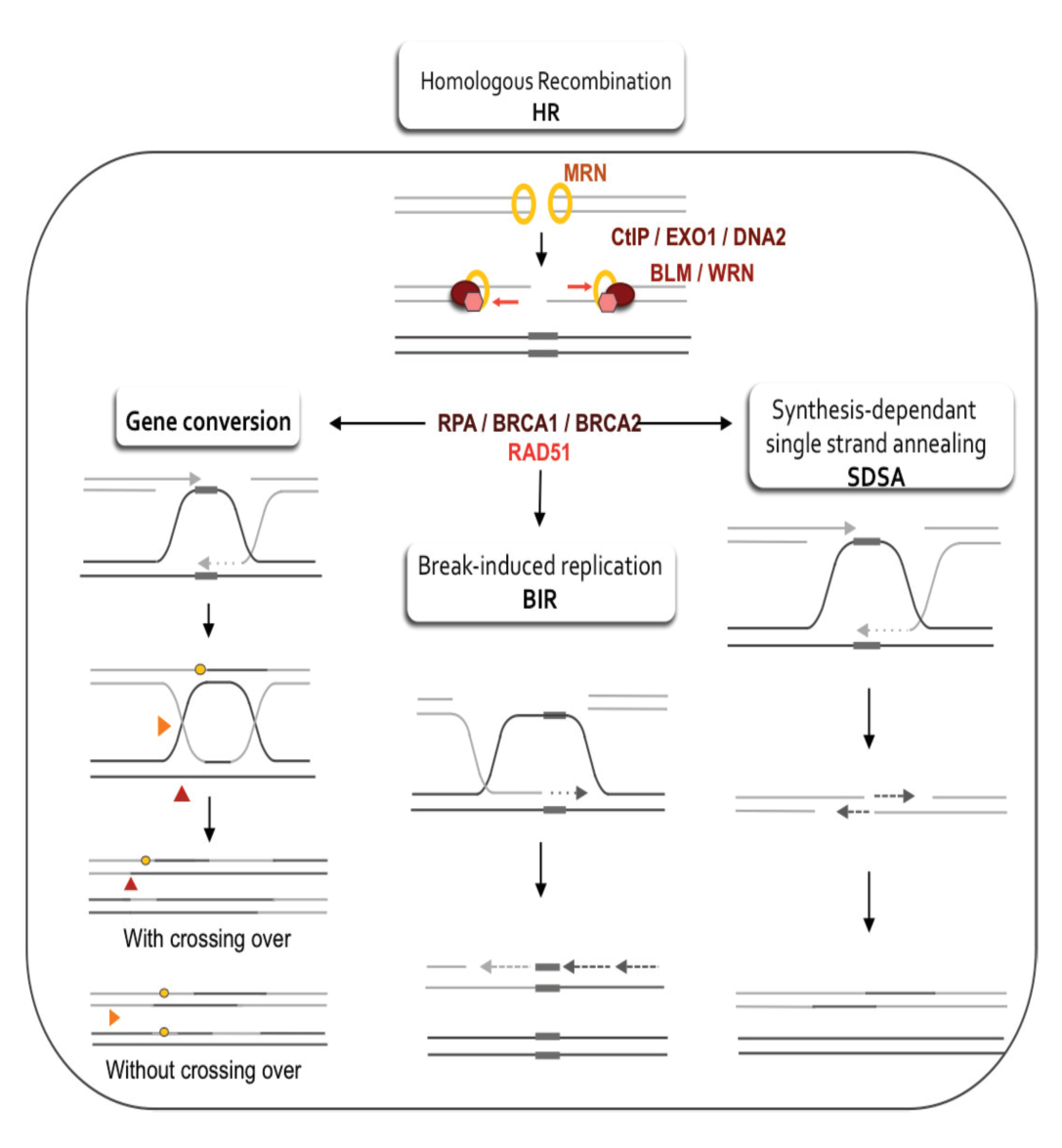
Homologous recombination
Homologous recombination comprises unwinding of the damaged DNA helix and attack of the damaged strands into a homologous DNA duplex molecule that may be the sister chromatid, homologous chromosome. For example, a vector or viral genome with homology in genomic DNA. Using the homologous strand as template, a DNA double-strand break takes place by homologous recombination replication. Homologous recombination therefore needs wide regions of DNA homology in another DNA duplex, and no loss of genetic information usually results.
Many genes and proteins involved in homologous recombination have been identified, mainly through yeast genetics and using mutants that are hypersensitive to ionizing radiation. These proteins are included in the products of the RAD51, RAD52, RAD54, RAD55, RAD57, RAD50, MRE11 and XRS2 genes. And most of these homologues have been cloned from mammals.
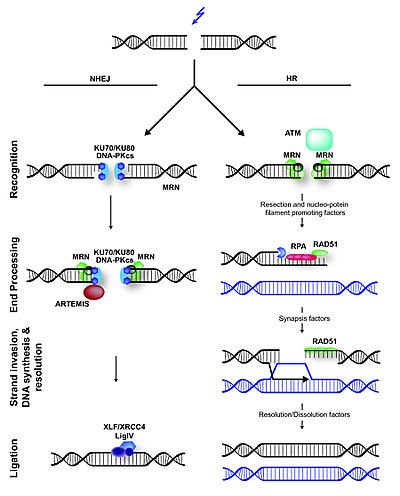
Non-homologous End joinig
In contrast to homologous recombination, nonhomologous end joining requires no homology with a second DNA duplex in contrast to homologous recombination. In mammals and in yeast, the first protein to recognize a DNA double-strand break seems to be Ku, a heterodimer of related ∼70 kDa and ∼80 kDa polypeptides. These polypeptides that together bind to the ends of broken DNA. It is not yet clear whether one Ku heterodimer is adequated to bind to the two ends at a break or whether one or more heterodimers bind to each end.
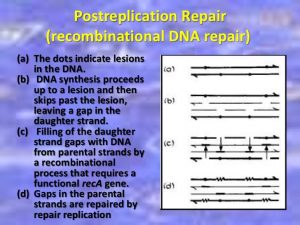
Post replication recombinant repair
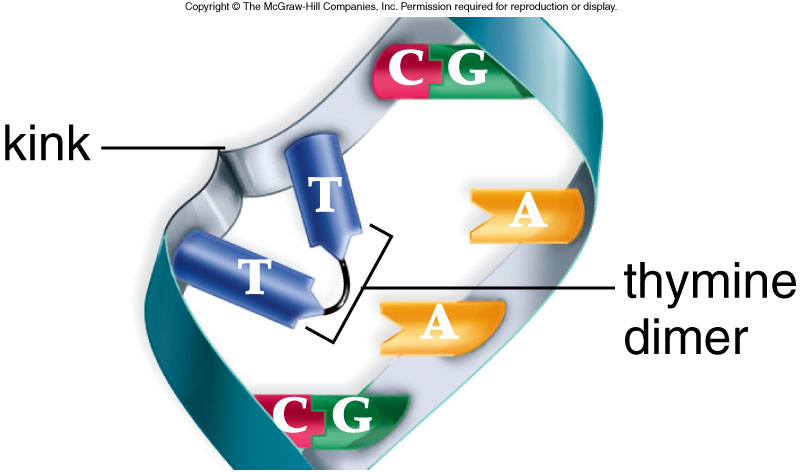
When DNA polymerase III encounters certain damage in E.coli, such as thymine dimers,it can not proceed. Instead the polymerase stops DNA synthesis and, leaving a gap, skip down the DNA to resume replication as far as eight hundred or more bases away. If allowed to remain, this gap is the result of deficiencies and broken DNA. Since part of one strand is absent and the other has been damaged so there seems to be no effective template to replicate new DNA.
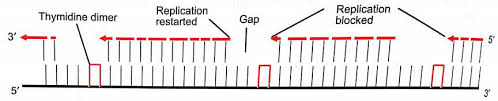
A replication fork creates 2 DNA duplexes. Thus, an undamaged copy of the region with lesion exists on the other daughter duplex. A group of enzymes, with one specified by the recA locus repairs the gap. Since the repair takes place at a gap created by failure of DNA replication, is called post replicative repair. This repair process that uses homologous DNA. It uses polymerases that can replicate these lesions.
This type of repair has been discovered in bacteria and mammalian cells.
Mechanisms for such repairs to the plant have not yet been described, although we know that no special efforts have been made.
In according to Rupp and Howard-Flanders (1968), the replicated complex jumps over that area in the parental strand of DNA in which the damage is localized, if the damage in the DNA is alive at the beginning of replication. As a result, UV-irradiated DNA immediately after replication consists of fragments the lengths of which band together with the distances between neighboring thymine dimers in the UV-damaged strand. Thus, the replication complex leaves gaps in the daughter strand opposed to the damaged area.
The repair of DNA is crucial to the development of every organism. Organisms have emanated many biochemical pathways for detecting and repairing DNA damage with high fidelity. Failure to repair DNA with high fidelity leads to a high conversion frequency. This is correlated with a cancer in humans. The type of DNA damage usually indicates the type of DNA repair pathway used by the cell.
This process was first familiar to use ultraviolet (UV)-irradiation as a source of DNA damage. While there are various types of UV-induced lesions (i.e., pyrimidine dimers), most are removed by the nuc1eotide-excision repair (NER) and photoreactivation repair (PR) pathways.
However, when a replication fork encounters one of these noncoding lesions (not removed by NER or PR), a special type of recombinational repair pathway is available to repair the DNA damage. The RecF pathway of recombinational DNA repair conducts on gapped DNA substrates that probably arise after the replication fork has partially replicated past a noncoding lesion in the template DNA.
Thus the damage is now in a DNA duplex and can be promoted either NER or PR. Recently PRR has been compacted into a broader paradigm that explains the coordination of DNA repair, DNA replication, and recombination in the cello This paradigm, called CPR (coordinated processing of damaged replication forks), combines many other aspects of DNA metabolism and cell division.
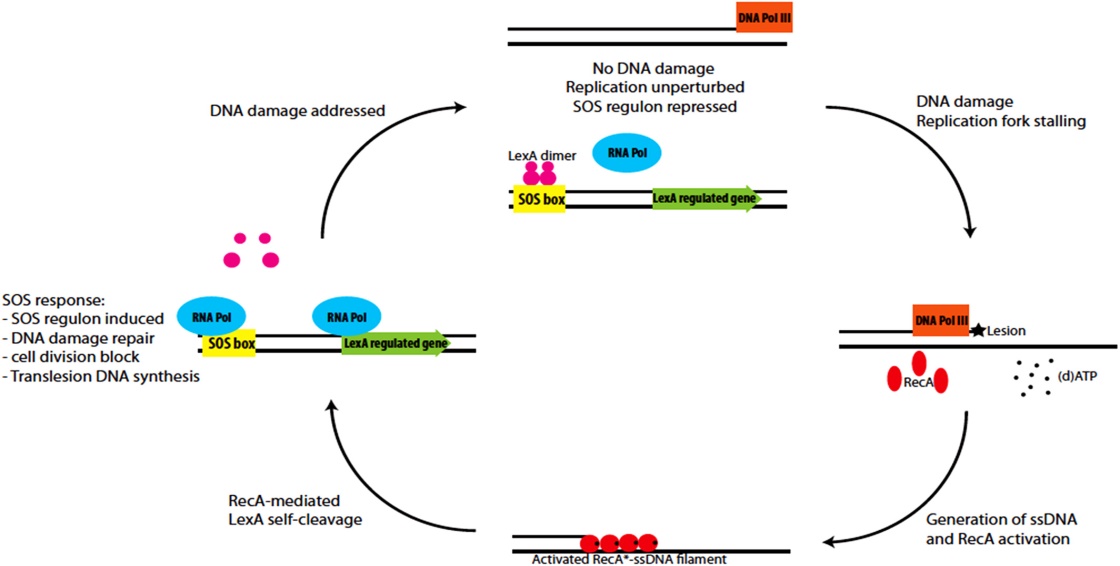
Sos repair
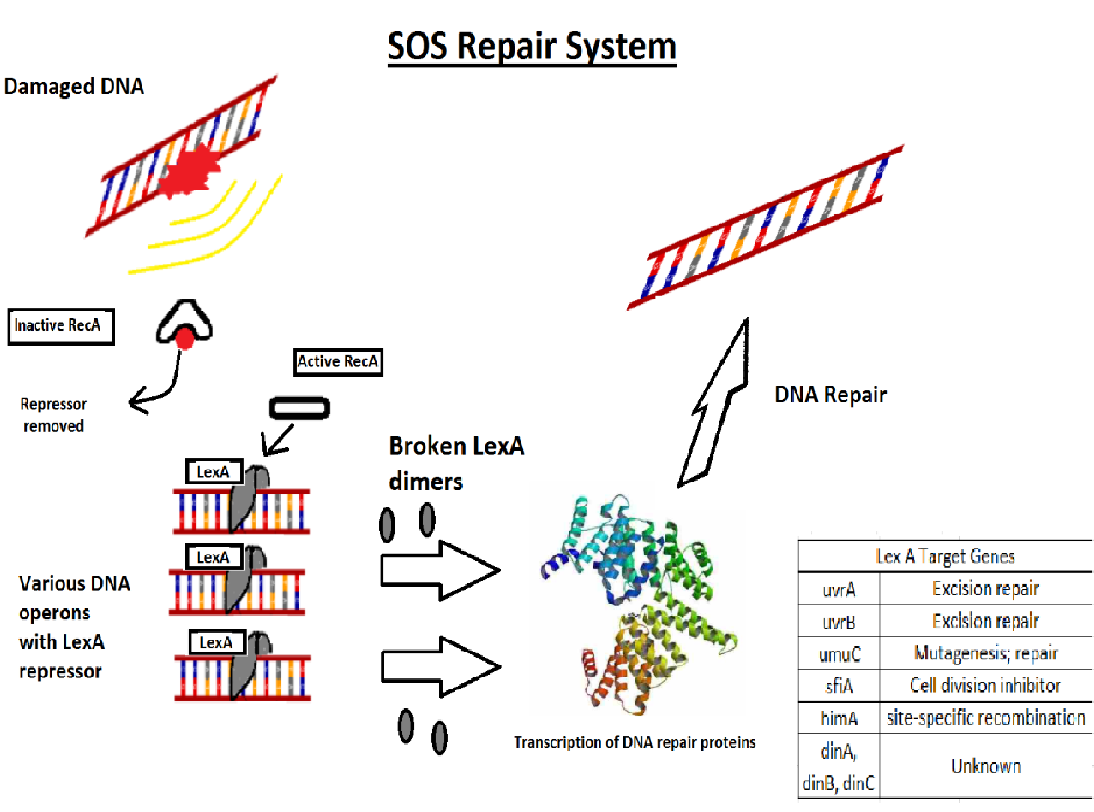
The SOS response may play a important role in persister cells generation , population-wide tolerance, and shielding. The SOS pathway is an invisible DNA damage repair system that is also important for bacterial adaptation, pathogenesis, and diversification. Several SOS regulators such LexA and RecA, some other stressors and stress responses can control SOS factors.
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka




