Transposons were first discovered in corn (maize) during the 1940s and ’50s by American scientist Barbara McClintock, whose work won her the Nobel Prize for Physiology or Medicine in 1983.
The colourful pattern on maize ears have an important scientific significance. Modern research have shown that the stripes and spots on maize karnels are the result of a genetic phenomenon called transposition. Within the maize indeed within the genome of most organisms geniticists found DNA sequences that can move from one position to another. These transposable elements or transposons constitute an appreciable fraction of the genome. elements (TEs), also known as “jumping genes,” are DNA sequences that move from one location on the genome to another. These elements were first identified more than 50 years ago by geneticist Barbara McClintock of Cold Spring Harbor Laboratory in New York. Biologists were initially skeptical of McClintock’s discovery. Over the next several decades, however, it became apparent that not only do TEs “jump,” but they are also found in almost all organisms (both prokaryotes and eukaryotes) and typically in large numbers.
TYPES OF TRANSPOSON
Since McClintock’s discovery, three basic types of transposons have been identified. These include class II transposons, miniature inverted-repeat transposable elements (MITEs, class III transposons), and retrotransposons (class 1transposons).
Transposable Elements In Bacteria
Although transposable elements were originally discovered ineukaryotes, bacterial transposons were the first to be studied at the molecular level. There are three main types: the insertion sequences, or IS elements, the composite transposons, and the Tn3-like elements. These three types of transposons differ in size and structure. The IS elements are the simplest, containing only genes that encode proteins involved intransposition. The composite transposons and Tn3-like elements are more complex, containing some genes that encode products unrelated to the transposition process.
IS ELEMENTS
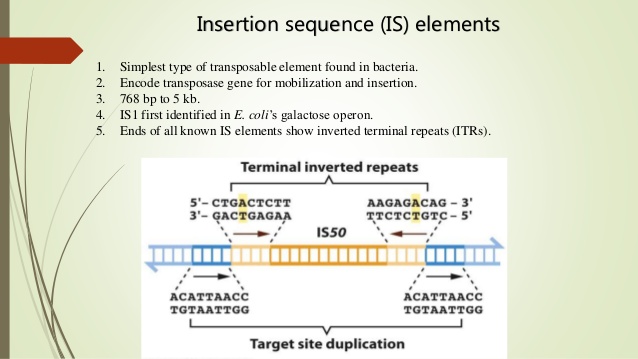
The simplest bacterial transposons are the insertion sequences, or IS elements, so named because they can insert at many different sites in bacterial chromosomes and plasmids. IS elements were first detected in certain lac mutations of E. coli. These mutations had the unusual property of reverting to wild-type at a high rate. Molecular analyses revealed that these unstable mutations possessed extra DNA in or near the lac genes. When DNA from the wild-type revertants of these mutations was compared with that from the mutations themselves, it was found that the extra DNA had been lost. Thus, these genetically unstable mutations were caused by DNA sequences that had inserted into E. coli genes, and reversion to wild-type was caused by excision of these sequences. Similar insertion sequences have been found in many other bacterial species.
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
IS elements are compactly organized. Typically, they consist of fewer than 2500 nucleotide pairs and contain only genes whose products are involved in promoting or regulating transposition. Many distinct types of IS elements have been identified.The smallest, IS1, is 768 nucleotide pairs long. Each type of IS element is demarcated by short identical, or nearly identical, sequences at its ends . Because these terminal sequences are always in inverted orientation with respect to each other, they are called terminal inverted repeats. Their lengths range from 9 to 40 nucleotide pairs. Terminal inverted repeats are characteristic of most—but not all types of transposons. When nucleotides in these repeats are mutated, the transposon usually loses its ability to move. These mutations therefore demonstrate that terminal inverted repeats play an important role in the transposition process.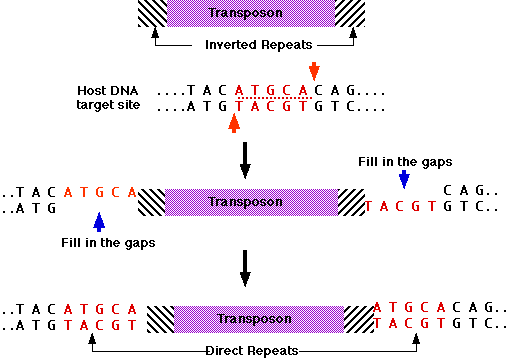
IS elements usually encode a protein, the transposase, that is needed for transposition. The transposase binds at or near the ends of the element and then cuts both strands of the DNA. This cleavage excises the element from the chromosome or plasmid, so that it can be
inserted at a new position in the same or a different DNA molecule. IS elements are therefore cut-and-paste transposons. When IS elements insert into chromosomes or plasmids, they create a duplication of part of the DNA sequence at the site of the insertion. One copy of the duplication is located on each side of the element. These short (2 to 13 nucleotide pairs), directly repeated sequences, called target site duplications, arise from staggered cleavage of the double-stranded DNA molecule. A bacterial chromosome may contain several copies of a particular type of IS element. For example, 6 to 10 copies of IS1 are found in the E.coli chromosome. Plasmids may also contain IS elements.
Cut-and-Paste Transposons in Eukaryotes
Geneticists have found many different types of transposons in eukaryotes. These elements vary in size, structure, and behavior. Some are abundant in the genome, others rare. In the following sections, we discuss eukaryotic transposon that move by a cut-and-paste mechanism. This element has inverted repeats at the terminal and create target site duplications when it inserts into DNA molecules. It encodes a transposase that catalyzes the movement of the element from one position to another.
Ac AND Ds ELEMENTS IN MAIZE
The Ac and Ds elements in maize were discovered by the American scientist Barbara McClintock. Through genetic analysis, McClintock showed that the activities of these elements are responsible for the striping and spotting of maize kernels. Many years later, Nina Federoff, Joachim Messing, Peter Starlinger, Heinz Saedler, Susan Wessler, and their colleagues isolated the elements and determined their molecular structure. McClintock discovered the Ac and Ds elements by studying chromosome breakage. She used genetic markers that controlled the color of maize kernels to detect the breakage events. When a particular marker was lost, McClintock inferred that the chromosome segment on which it was located had also been lost, an indication that a breakage event had occurred. The loss of a marker was detected by a change in the color of the aleurone, the outermost layer of the triploid endosperm of maize kernels. In one set of experiments, the genetic marker that McClintock followed was an allele of the C locus on the short arm of chromosome 9. Because this allele, CI, is a dominant inhibitor of aleurone coloration, any kernel possessing it is colorless. McClintock fertilized CC ears with pollen from CICI tassels, producing kernels in which the endosperm was CICC. (The triploid endosperm receives two alleles from the female parent and one from the male parent) Although McClintock found that most of these kernels were colorless, as expected, some showed patches of brownish-purple pigment. McClintock guessed that in suchmosaics, the inhibitory CI allele had been lost sometime during endosperm development, leading to a clone of tissue that was able to make pigment. The genotype in such a clone would be -CC, where the dash indicates the missing CI allele.
The mechanism that McClintock proposed to explain the loss of the CI allele is diagrammed . A break at the site labeled by the arrow detaches a segment of the chromosome from its centromere, creating an acentric fragment. Such a fragment tends to be lost during cell division; thus, all the descendants of this cell will lack part of the paternally derived chromosome. Because the lost fragment carries the CI allele, none of the cells in this clone is inhibited from forming pigment. If any of them produces a part of the aleurone, a patch of purple tissue will appear, creating a mosaic kernel similar to the one.
McClintock found that the breakage responsible for these mosaic kernels occurred at a particular site on chromosome 9. She named the factor that produced these breaks Ds, for Dissociation. However, by itself, this factor was unable to induce chromosome breakage. In fact, McClintock found that Ds had to be stimulated by another factor, called Ac, for Activator. The Ac factor was present in some maize stocks but absent in others. When different stocks were crossed, Ac could be combined with Ds to create the condition that led to chromosome breakage. This two-factor Ac/Ds system provided an explanation for the genetic instability that McClintock had observed on chromosome 9. Additional experiments demonstrated that this was only one of many instabilities present in the maize genome. McClintock found other instances of breakage at different sites on chromosome 9 and also on other chromosomes. Because breakage at these sites depended on activation by Ac, she concluded that Ds factors were also involved. To explain all these observations, McClintock proposed that Ds could exist at many different sites in the genome and that it could move from one site to another.
This explanation has been borne out by subsequent analyses. The Ac and Ds elements belong to a family of transposons. These elements are structurally related to each other and can insert at many different sites on the chromosomes. Multiple copies of the Ac and Ds elements are often present in the maize genome. Through genetic analysis, McClintock demonstrated that both Ac and Ds can move. When one of these elements inserts in or near a gene, McClintock found that the gene’s function is altered—sometimes completely abolished. Thus, Ac and Ds can induce mutations by inserting into genes. To emphasize this effect on gene expression, McClintock called the Ac and Ds transposons controlling elements.
DNA sequencing has shown that Ac elements consist of 4563 nucleotide pairs bounded by inverted repeats that are 11 nucleotide pairs long ; these terminal inverted repeats are essential for transposition. Each Ac element is also flanked by direct repeats 8 nucleotide pairs long. Because the direct repeats are created at the time the element is inserted into the chromosome, they are target site duplications, not integral parts of the element.
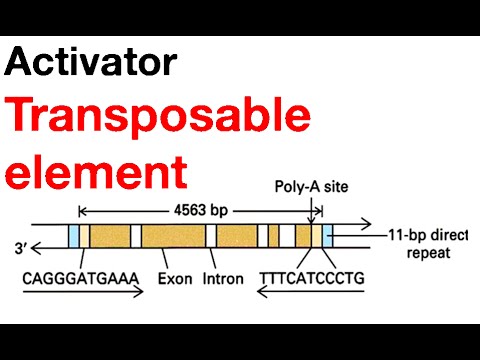
Unlike Ac, Ds elements are structurally heterogeneous. They all possess the same inverted terminal repeats as Ac elements, demonstrating that they belong to the same transposon family, but their internal sequences vary. Some Ds elements appear to havebeen derived from Ac elements by the loss of internal sequences . The deletions in these elements may have been caused by incomplete DNA synthesis during replication or transposition. Other Ds elements contain non-Ac DNA between their inverted terminal repeats These unusual members of the Ac/Ds family are called aberrant Ds elements. A third class of Ds elements is characterized by a peculiar piggy backing arrangement one Ds element is inserted into another but in an inverted orientation. These so-called double Ds elements appear to have been responsible for the chromosome breakage that McClintock observed in her experiments.

The activities of the Ac/Ds elements—excision and transposition, and all their genetic correlates, including mutation and chromosome breakage—are caused by a transposase encoded by the Ac elements. The Ac transposase interacts with sequences at or near the ends of Ac and Ds elements, catalyzing their movement. Deletions or mutations in the gene that encodes the transposase abolish this catalytic function. Thus Ds elements, which have such lesions, can not activate themselves. However, they can be activated if a transposase producing Ac element is present somewhere in the genome. The transposase made by this element can diffuse through the nucleus, bind to a Ds element and activate it. The Ac transposase is, therefore, a trans-acting protein. Transposons related to the Ac/Ds elements have been found in other species including animals. Perhaps the best-studied of these elements is one called hobo, whimsically named for its ability to transpose. The hobo element
is found in some species of Drosophila.
The Ac-Ds system in Maize
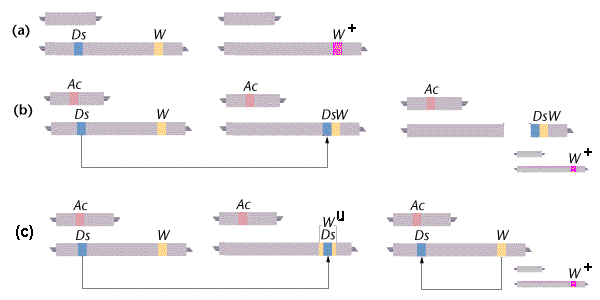
In the presence of an Activator (Ac) element, a Dissociator (Ds) element can cause chromosome breaks. The genetic marker for such breaks in this example is expression at the W locus: the dominant W allele produces a mutant White phenotype in the individual seeds (kernels), and the recessive W+ allele produces a wild-type dark phenotype.
(a) In the absence of an Ac element, Ds is not transposable and chromosome breaks do not occur. In a W / W+ heterozygote, the W allele is expressed as expected and the plant is white.
(b) In the presence of an Ac element, the Ds element can be transposed (“jump“) elsewhere in the chromosome. This transposition can cause a chromosome break at the point of insertion, so that the Ds element and any downstream loci are lost. In a W / W+heterozygote, loss of the W allele allows expression of the alternative W+ allele on the other chromosome, producing an unexpected dark phenotype. [The second chromosome set is the same as in (a), shown reduced].
(c) Ds transposition can also cause the element to jump into the middle of the W allele, disrupting its normal function and again allowing expression of the alternative W+ allele. The seeds are then a variegated, mosaic pattern of cell patches, with a background due to W and a superimposed pattern of or W+ spots.
However, the disruption is unstable (Wu), as Ds may subsequently jump back out of Wu, restoring function of W. [The second chromosome set is again the same as in (a), shown reduced]
The Ac – Ds system in maize (Zea): genetics of “jumping genes“
Sources:
- https://www.nature.com/scitable/topicpage/Barbara-McClintock-and-the-Discovery-of-Jumping-34083
- TEXTBOOK: Principles of Genetics by SNUSTED and SIMMONS.
Revised by Md Siddiq Hasan on 29/07/20
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka




