Genes are the carriers of all the information of an organism. These information are needed for different protein synthesis at different time. So expression of gene can effect an organism’s characteristics, health and so on.
As gene expression is a very important feature for any organism as well as for the prokaryotes, the mechanism which controls this feature is also extremely important for an organism. We can call this mechanism as ‘Regulation of Gene Expression’.
INDUCTION
There are some enzymes, synthesis of which depends upon the presence of a specific substrate. These enzymes are called inducible enzymes and the genetic system responsible for the synthesis of such an enzyme is known as inducible system.
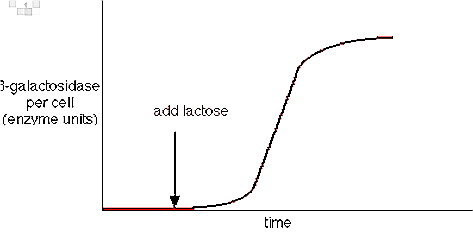
For example, in Escherichia coli there is an enzyme called ß-galactosidase which is important for hydrolysis of lactose into glucose and galactose. It was known since early 1960s.
Lactose => Glucose + Galactose
If the substrate lactose is not supplied, the presence of enzyme ß-galactosidase becomes very less to be detected. But with the addition of lactose the mentioned enzyme increases as much as 10,000 fold. The proportion of enzyme in E. coli cells again falls down if the substrate lactose is removed.
REPRESSION
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
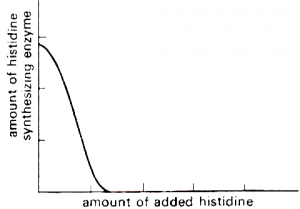
There is a case known as repression in which synthesis of enzyme is restricted by adding end product of the bio-synthetic pathway. This type of enzymes are known as repressible enzyme and the genetic system responsible for it is called Repressible system. For instance, if no amino acid is supplied, E. coli cells can synthesize all enzymes which are needed for production of different amino acids. If histidine is added, production of histidine will decrease severely.
Transcriptional regulation
Operon model
This model was first proposed by F. Jacob and J. Monod. In this proposal, the model assumed a unit of coordinated control of protein synthesis. The mentioned unit was called ‘Operon‘.
An operon is consisted of-
- An Operator Gene
- Structural proteins
The operator gene controls the activity of the structural genes. The secondly mentioned genes are characterized to participate in the synthesis of proteins.
Structural genes synthesize mRNA, which is controlled by operator gene. The mechanism of regulation of protein synthesis using operon model can be pictured through different type sof operons like lac operon, arabionose operon, tryptophan operon.
Lac operon
Like other operons, lac operon consists of one operator and some structural genes.
The operator consists of 26 base pairs. Three structural genes are present in this operon. They are:
-
- Gene lac Z
- Gene lac Y
- Gene lac A
Gene lac Z
It is consisted of 3063 base pairs. The job of this structural gene is to code for the enzyme ß-galactosidase. This enzyme remains active as a tetramer and works to break lactose into glucose and galactose. The operator of lac operon is a part of this structural gene.
Gene lac Y
It consists of 800 base pairs. It codes for Beta-galactose permease. This product is a membrane bound protein. It helps in transportation of metabolites.
Gene lac A
800 base pairs are involved in this gene. It codes for Beta-galactose transacetylase enzyme. This enzyme transfers an acetyl group from acetyl-CoA to Beta-galactosides.
If mutation occurs in lac Z and lac Y, lactose is not utilized by the cell.
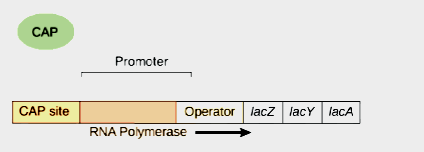
In lac operon, by the activity of gene lac I, repressor is synthesized. Gene lac I is known as regulator gene. The repressor synthesized by it attaches on operator gene and stops the synthesis of mRNA. If lactose is added to the system, repressor becomes inactive . As a result, mRNA synthesis takes place.
A promoter gene controls the activity of regulator gene. This promoter gene is different from the promoter gene for lac operon. This is situated at the 5′ end.
Promoter gene for lac operon
The actual site for transcription is promoter gene. On the other hand operator gene is the site of action of repressor. Promoter gene lies upstream to the repressor. RNA polymerase binds to the promoter site. The movement of RNA polymerase starts from here.
CAP or CGA site
CAP stands for ‘catabolite activator protein’ or ‘Cyclic AMP protein’. And CGA stands for ‘catabolite gene activator’. This is a protein which binds to the mentioned site. This site is situated upstream of the promoter. It consists of 16 base pairs. The protein which binds to the site interacts with DNA. It has a positive control. When cyclic AMP molecules activates cga protein , then RNA polymerase is allowed to bind. A protein which binds with DNA and assists in transcription is called apoinducer. So CAP protein is called apoinducer.
Tryptophan operon
An operator and five structural genes are involved in the synthesis of tryptophan. And these makes the operon called tryptophan operon. These code for three enzymes needed for tryptophan synthesis.
Promoter site
Transcription starts at this site. It is situated upstream of the structural genes.
Repressor
Gene trp R codes for repressor. The repressor is not linked with the operon. This repressor generally remains inactive as it has low affinity to the operator. It becomes active when it is associated with tryptophan. Because tryptophan works as co-repressor. Thus the synthesis of tryptophan is partly self-regulatory. The work of tryptophan as co-repressor is to stop further synthesis of tryptophan.
Tryptophan repressor controls three set of genes.
Trp repressor controls three sets of unlinked genes. These are:
- trp EDBCA
- aroH
- trp R
trp EBCDA controls the enzymes which are involved in synthesis of tryptophan. aroH gene codes for an enzyme needed for a common pathway of aromatic acid biosynthesis. trp R is regulator gene which is repressed by it’s own gene product.
An operator gene is located at each of these genes. It may be present within the promoter or downstream or upstream of the promoter.
Thus negative control is seen in tryptophan operon.
Arabinose operon
This is an operon which has a positive control over gene expression. This has three structural genes named as O, B and A. O is operator site. The initiating site is named as ‘i’. There is another gene named as ‘C’. It is indicated for regulator gene.
The regulator gene C is comparable to the regulator gene of lac operon. The C gene gives rise to a protein indicated as P1. This protein works as repressor in the absence of arabinose. In the presence of arabinose, P1 is converted into another protein named mentioned as P2. This protein works as a stimulator in transcription. Thus transcription occurs with the help pf the operon.
Negative and Positive Controls of Transcription
Lactose operon and tryptophan operon have the systems of regulation which are negative. Because the operon is generally active but it is kept inactive by the regulator gene. However, itbis described that with the help of CGA protein , positive control is also maintained by lac operon.
The regulator gene stimulates the production of enzyme in a positive control by operon. This is seen in arabinose operon.
Thus these operons both negative and positive, regulates the expression of gene in prokaryotes.
CONCLUSION
These mechanisms responsible for gene expression can be said as the ‘mechanisms for existing life on earth‘ . Because if this mechanisms fail , protein synthesis will not occur. And all the organism’s life is depended on these different kinds of protein. So synthesis of these are necessary for all the life forms .
Thus regulation of gene expressions give certainty of their life to the organisms when that expression is needed.
Reference:
- GENETICS Classical to Modern by P.K. Gupta
- www.wikipedia.org
Revised by Md. Siddiq Hasan on 28/07/20
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka





Your article helped me a lot, is there any more related content? Thanks!