The four bases in DNA – A, T, G, and C are sufficient to specify the 20 amino acids in proteins because each codon is three bases in length. Each sequence of three adjacent bases in mRNA is a codon that specifies a particular amino acid (or chain termination). The genetic code is the list of all codons and the amino acid that each one encodes. Before the genetic code was determined experimentally, it was reasoned that if all codons were assumed to have the same number of bases, then each codon would have to contain at least three bases.
Why does each codon have 3 bases?
Codons having only one base at a time will make only 4 types which are insufficient for making 20 amino acids. Again codons consisting of pairs of bases would be insufficient, because four bases can form only 42 = 16 pairs; triplets of bases would suffice, because four bases can form 43 = 64 triplets.
In fact, the genetic code is a triple code, and all 64 possible codons carry information of some sort. Most amino acids are encoded by more than one codon. Furthermore, in the translation of mRNA molecules, the codons do not overlap but are used sequentially.
Note that
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
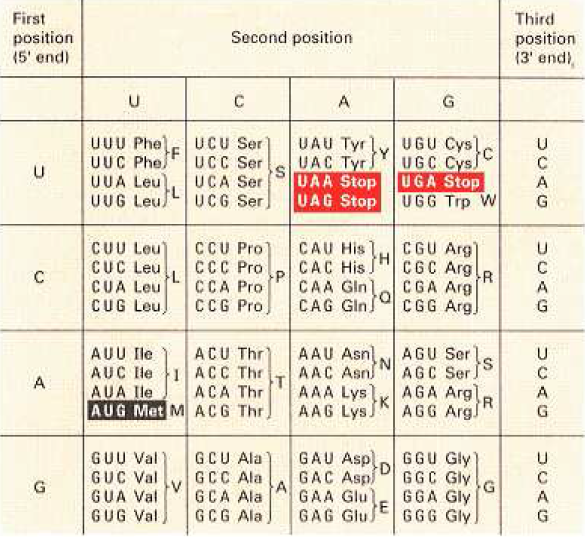
- 61 codons out of 64 specify amino acids.
- 4 codons are signal codons. One (AUG) is a start codon and also specifies Methionine and the other 3 are stop codons and do not specify amino acids.
Understanding Mutation in Genes
Mutations that change the nucleotide sequence of a gene may differ in their consequences, and a special terminology is used to describe them.
Missen mutation: A missens mutation results in the replacement of one amino acid by another. For example, the change from GAG to GTG in the DNA of the gene for b-globin results in the replacement of glutamic acid by valine in the sickle-cell hemoglobin molecule; the mutation is therefore a missense mutation.
Silent mutation: A silent mutation is one that does not change the amino acid sequence. Silent mutations often result from changes in the third codon position (meaning third sequence in codon is non-significant); for example, a mutation that changes an AAA codon into an AAG codon is silent because both codons specify lysine (see the chart above).
Nonsense mutation: A most interesting class of mutations consists of changes that convert a codon that specifies an amino acid into a chain-terminating codon. A mutation of this type is called a nonsense mutation, and it results in premature termination of the polypeptide chain. An example of a nonsense mutation is found in the b-globin gene, in which a mutation from AAG to UAG in the seventeenth codon results in a truncated polypeptide only 16 amino acids in length. This mutation is one of several types associated with the disease b-thalassemia.
The silent mutation has baffled the scientists for quite some time and made them come out with hypothesis to explain the puzzle of it along with some other phenomena.
- The identity of the third base of a codon is often unimportant.Example: Silent mutation.
- Codon is highly redundant (Although each codon is specific for only one amino acid (or one stop signal), the genetic code is described as degenerate, or redundant, because a single amino acid may be coded for by more than one codon.)For example, Serine (Ser) can be coded by any of 4 codons: UCU, UCC, UCA and UCG.
- The number of distinct tRNA molecules that have been isolated from a single organism is less than the number of codons; so, the anticodons of some tRNA molecules must be able to pair with more than one codon.(There are 61 codons that code for amino acids. And amino acids are carried by tRNAs. So, earlier scientists presumed that there should be 61 tRNAs for 61 codons. But later it was found that the tRNAs present in a cell are less in number. So, there must be multiple codons that can pair with single tRNA molecule.)
Wobble hypothesis
To account for these observations, the wobble concept was advanced in 1966 by Francis Crick. He proposed that the first two bases in a codon form base pairs with the tRNA anticodon according to the usual rules (A-U and G-C) but that the base at the 5′ end of the anticodon is less spatially constrained than the first two and can form hydrogen bonds with more than one base at the 3′ end of the codon.
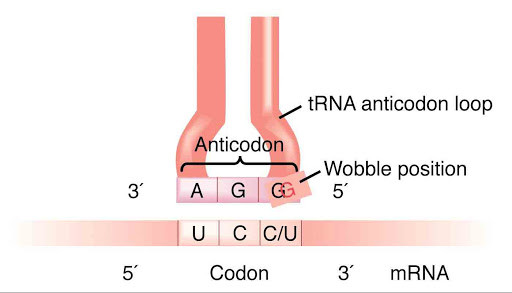
For example, in the image above, the 5′ end base of anticodon is G which can attach with any base between U and C of 3′ end of the mRNA codon. This proves the concept of multiple codons for one tRNA otherwise known as wobble hypothesis.
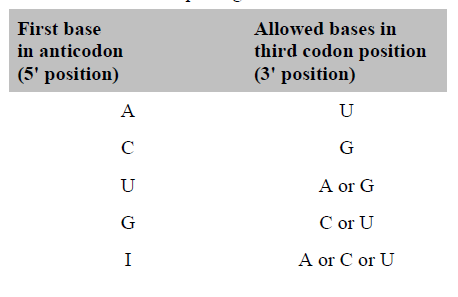 He suggested the pairing rules given in the table. Evidence has confirmed the wobble concept and indicates that the pairings given in the table are largely true for E. coli. On the other hand, analysis of tRNAs in the yeast Saccharomyces cerevisiae has indicated that wobble is more restricted in yeast than in E. coli. The table summarizes the wobble rules for E. coli and yeast. A total of 46 tRNAs are needed to decode mRNA molecules in yeast. As noted, there are no tRNAs corresponding to the stop codons.
He suggested the pairing rules given in the table. Evidence has confirmed the wobble concept and indicates that the pairings given in the table are largely true for E. coli. On the other hand, analysis of tRNAs in the yeast Saccharomyces cerevisiae has indicated that wobble is more restricted in yeast than in E. coli. The table summarizes the wobble rules for E. coli and yeast. A total of 46 tRNAs are needed to decode mRNA molecules in yeast. As noted, there are no tRNAs corresponding to the stop codons.
Characteristics of Genetic Code
1. The genetic code consists of a sequence of nucleotides in DNA or RNA. There are four letters in the code, corresponding to the four bases—A, G, C, and U (T in DNA).
2. The genetic code is a triplet code. Each amino acid is encoded by a sequence of three consecutive nucleotides, called a codon.
3. The genetic code is degenerate—there are 64 codons but only 20 amino acids in proteins. Some codons are synonymous, specifying the same amino acid.
4. Isoaccepting tRNAs are tRNAs with different anticodons that accept the same amino acid; wobble allows the anticodon on one type of tRNA to pair with more than one type of codon on mRNA.
5. The code is generally non-overlapping; each nucleotide in an mRNA sequence belongs to a single reading frame.
6. The reading frame is set by an initiation codon, which is usually AUG.
7. When a reading frame has been set, codons are read as successive groups of three nucleotides.
8. Any one of three termination codons (UAA, UAG, and UGA) can signal the end of a protein; no amino acids are encoded by the termination codons.
9. The code is almost universal.
Source:
- Genetics by Heartl Jones
Revised by Md. Siddiq Hasan on 27/07/20
Get Free Netflix Now
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka