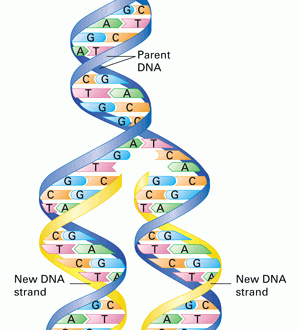
DNA replication in eukaryotes is a complex and unique process involving many enzymes and thousands of ORI at a single time. Unlike the prokaryotic DNA, it involves a linear mode of replication.
Why does linear DNA replication involve multiple origins at a time?
The large linear chromosomes in eukaryotic cell contain far too much DNA to be replicated speedily from a single origin. Eukaryotic replication proceeds at a rate ranging from 500 to 5000 nucleotides per minute at each replication fork (considerably slower than bacterial replication). Even at 5000 nucleotides per minute at each fork, DNA synthesis starting from a single origin would require 7 days to replicate a typical human chromosome consisting of 100 million base pairs of DNA. The replication of eukaryotic chromosomes actually occurs in a matter of minutes or hours, not days. This rate is possible because replication takes place simultaneously from thousands of origins.
Requirements
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
Although the process of replication includes many components, they can be combined into three major groups:
- A template consisting of single-stranded DNA,
- Raw materials (substrates) to be assembled into a new nucleotide strand (Deoxyribonucleoside triphosphates (dNTPs) – dATP, dCTP, dTTP, dGTP)
- Enzymes and other proteins that “read” the template and assemble the substrates into a DNA molecule.
Good to know
- Typical eukaryotic replicons are from 20,000 to 300,000 base pairs in length.
- The nucleotides that form the new strand following the template always come as Triphophates, not monophosphates, although monophosphate nucleoside is required for the growing chain (which is actually formed by breaking the triphosphate).
- The bond between one nucleotide to another is phosphodiester bond which basically adds the phosphate group coming nucleotide to the -OH group of preceding and releases a molecule of water.
Direction of Replication
(5′ to 3′)
In DNA synthesis, new nucleotides are joined one at a time to the 3′ end of the newly synthesized strand. Why?
DNA polymerases, the enzymes that synthesize DNA, can add nucleotides only to the 3′ end of the growing strand (not the 5′ end), so new DNA strands always elongate in the same 5′-to-3′ direction.
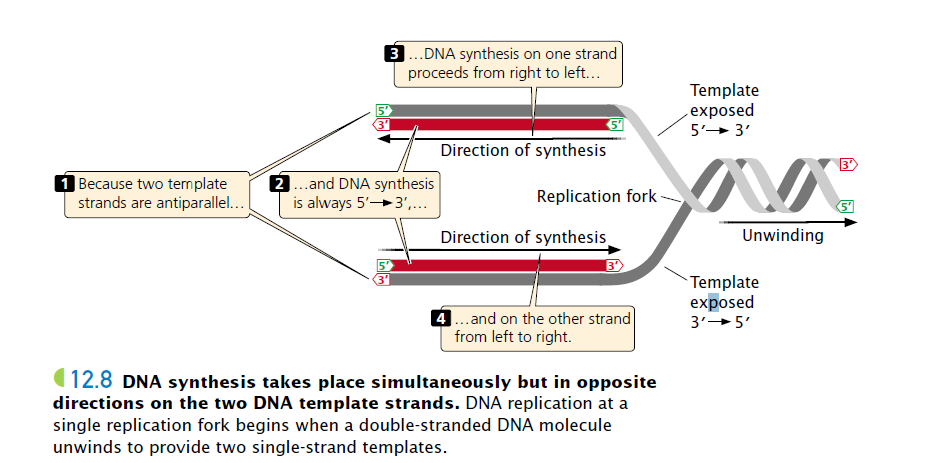
(Because the two single stranded DNA templates are antiparallel (meaning: one is from 5′ to 3′ and another is from 3′ to 5′) and strand elongation is always 5′ to 3′, if synthesis on one template proceeds from, say, right to left, then synthesis on the other template must proceed in the opposite direction, from left to right.)
Eukaryotic DNA Replication
(Step by step approach)
Before starting this part of the article, understand the followings:
- The replication starts at multiple origins. For convenience, we will talk about only one origin of replication.
- Replication fork proceeds in bidirection from the origin. We will consider only one direction here. The other fork will follow the same rules to replicate.
Remember, that the enzyme sets required for replication in an origin is 2 in number, as replication proceeds in both direction.
Unwinding
Because DNA synthesis requires a single stranded template, double-stranded DNA must be unwound before DNA synthesis can take place.
- DNA helicases break the hydrogen bonds that exist between the bases of the two nucleotide strands of a DNA molecule.
- Helicase moves from 5′ to 3′ direction.
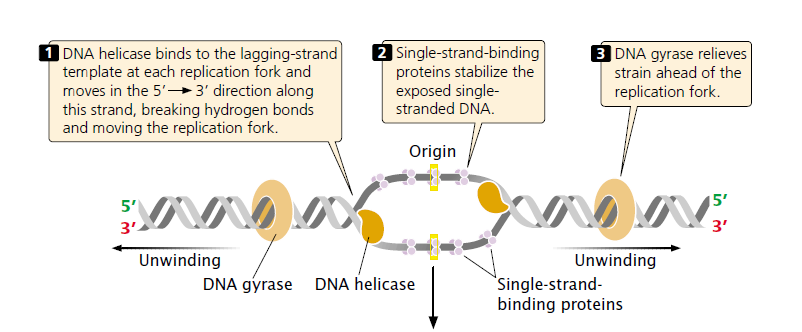
After DNA has been unwound by helicase, the single stranded nucleotide chains have a tendency to form hydrogen bonds and reanneal (stick back together).
- To stabilize the single-stranded DNA long enough for replication to take place, single-strand-binding (SSB) proteins attach tightly to the exposed single-stranded DNA.
- DNA gyrase enzyme prevents the strands from supercoiling again.
In eukaryotes, the functions of these both enzymes are performed by topoisomerases.
Adding Primers
|
(This is the step where the DNA replication actually begins. Here, the most important enzyme of DNA replication called DNA polymerase is needed. DNA polymerase attaches the nucleotides in the new chain one by one. But it has a limitation. In order to add new nucleotides it needs an already existing 3′ OH end. So in bare template or without the presence of something with a 3′ terminal (primer actually), DNA polymerase can not start replication) |
A primer is a short strand of single stranded RNA which has both 5′ and 3′ end. An enzyme called primase synthesizes primers to get DNA replication started.
- A primer is about 10-12 nucleotides long.
- provides a 3′-OH group to which DNA polymerase can attach DNA nucleotides.
- All DNA molecules initially have short RNA primers embedded within them; these primers are later removed and replaced by DNA nucleotides.
So, at the point where replication starts, primers are added to start the function of DNA polymerase enzymes.
Elongation
After DNA is unwound and a primer has been added, DNA polymerases elongate the growing chain by adding nucleotides one by one.
|
(Note that after adding the first nucleotide to the primer’s 3′ end, the added nucleotide makes another 3′ end of itself open. So, no problem arises to add the second nucleotide and this way the succession of polymerization continues.) (Secondly, the primer has another end 5′. As the chain is growing along the 3′ end, the direction of replication is from 5′ to 3′) |
The DNA polymerases
- synthesize in the 5′ to 3′ direction by adding nucleotides to a 3′-OH group;
- use dNTPs to synthesize new DNA;
- require a primer to initiate synthesis;
- catalyze the formation of a phosphodiester bond by joining the 5′ phosphate group of the incoming nucleotide to the 3′-OH group of the preceding nucleotide on the growing strand, cleaving off two phosphates in the process;
- produce newly synthesized strands that are complementary and antiparallel to the template strands;
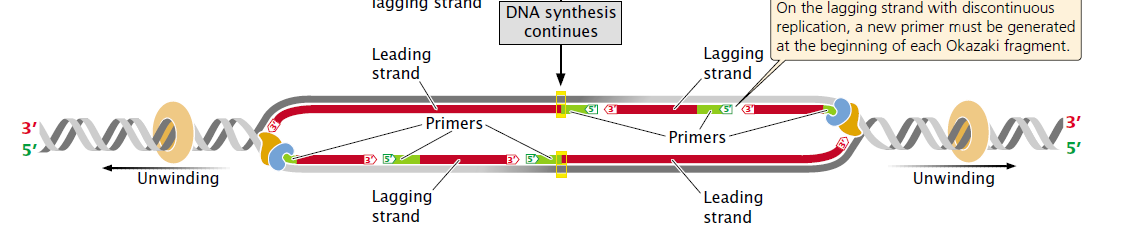
Leading strand (Lower red strand right to the ORI)
As the DNA unwinds, the template strand that is exposed in the 3′ to 5′ direction (the lower strand in the figure) allows the new strand to be synthesized continuously in the 5′ to 3′ direction. This new strand, which undergoes continuous replication, is called the leading strand.
| (It grows continuously because the direction of growing chain i.e adding nucleotides by DNA polymerase (left to right) is towards the direction of the replication fork moving right to the origin of replication.) |
Lagging strand (Upper red strand right to the ORI)
The other template strand is exposed in the 5′ to 3′ direction (the upper strand in the figure). After a short length of the DNA has been unwound, synthesis must proceed 5′ to 3′; that is, in the direction opposite that of unwinding.
Because only a short length of DNA needs to be unwound before synthesis on this strand gets started, the replication machinery soon runs out of template. By that time, more DNA has unwound, providing new template at the 5′ end of the new strand.
DNA synthesis must start anew at the replication fork and proceed in the direction opposite that of the movement of the fork until it runs into the previously replicated segment of DNA. This process is repeated again and again, so synthesis of this strand is in short, discontinuous bursts. The newly made strand that undergoes discontinuous replication is called the lagging strand.
The short lengths of DNA produced by discontinuous replication of the lagging strand are called Okazaki fragments, after Reiji Okazaki, who discovered them.
|
(Understanding lagging strand: এখানে অধারাবাহিকভাবে স্ট্র্যান্ড তৈরী হওয়ার প্রধান কারণটি হচ্ছেঃ ডিএনএ স্ট্র্যান্ড তৈরী হয় একদিকে কিন্তু রেপ্লিকেশন ফর্ক খোলে আরেকদিকে। যেহেতু রেপ্লিকেশন ফর্ক খোলার প্রায় সাথে সাথেই প্রাইমার তৈরী হয়ে পলিমারাইজেশন হতে থাকে অরিজিনের দিকে (৫ থেকে ৩ অবশ্যই), যতক্ষণে এই প্রাইমারের বামে নিউক্লিয়োটাইডগুলো যোগ হচ্ছে ততক্ষণে আবার ফর্কটি ডানে কিছুদূর এগিয়ে যায়। যেহেতু ডিএনএ পলিমারেজ শুধু প্রাইমারের ৩ মাথাতেই নিউক্লিয়োটাইড লাগাতে পারে, কিছুদূর দূরে খোলা ফর্কে আবার নতুন একটি প্রাইমার তৈরী হয়, সেখানে আবার ৩ মাথাতে নিউক্লিয়োটাইড যোগ হয়ে হয়ে আগের প্রাইমারের ৫ মাথার কাছাকাছি চলে আসে। কিন্তু যোগ হয় না। এসব খন্ডের কারণে একে ডিসকন্টিনিউয়াস স্ট্র্যান্ড বলা হয়। এবং খন্ডগুলোকে বলা হয় ওকাজাকি খন্ড। এদিকে ততক্ষণে আবার রেপ্লিকেশন ফর্কটি এগিয়ে যায় এবং তৃতীয় আরেকটিই প্রাইমার তৈরী হয়। এভাবেই রিপিটেডলি প্রক্রিয়াটি চলতে থাকে।) |
DNA ligase
The DNA polymerase I replaces the Primers (RNA nuclotides) and replaces them with dNTPs. But still after removing the ligases, nicks remain present between each two Okazaki fragments. And here DNA ligase comes into play. It attaches the okazaki fragments and make the strands continuous.
Termination
In some DNA molecules, replication is terminated whenever two replication forks meet. In others, specific termination sequences block further replication.
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka