The polymerase chain reaction, generally known as PCR, is used in almost every application of gene technology. It is a method for rapid production of a very large number of copies of a particular fragment of DNA. Virtually unlimited quantities of a length of DNA can be produced from the smallest quantity of DNA (even one molecule). Figure A shows the steps involved in PCR.
What is PCR?
PCR is a technique used in the lab to make millions of copies of a particular section of DNA. It was first developed in the 1983.
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
- The polymerase chain reaction (PCR) was originally developed in 1983 by the American biochemist Kary Mullis. He was awarded the Nobel Prize in Chemistry in 1993 for his pioneering work.
- PCR is used in molecular biology to make many copies of (amplify) small sections of DNA or a gene.
- Using PCR it is possible to generate thousands to millions of copies of a particular section of DNA from a very small amount of DNA.
- PCR is a common tool used in medical and biological research labs. It is used in the early stages of processing DNA for sequencing? , for detecting the presence or absence of a gene to help identify pathogens ?during infection, and when generating forensic DNA profiles from tiny samples of DNA.
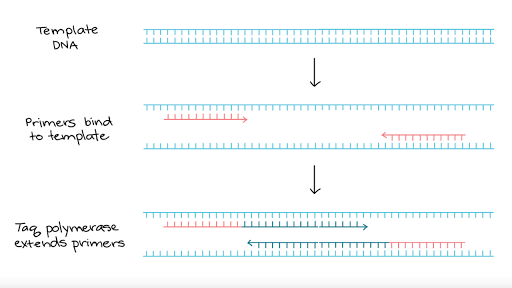
One cycle of PCR
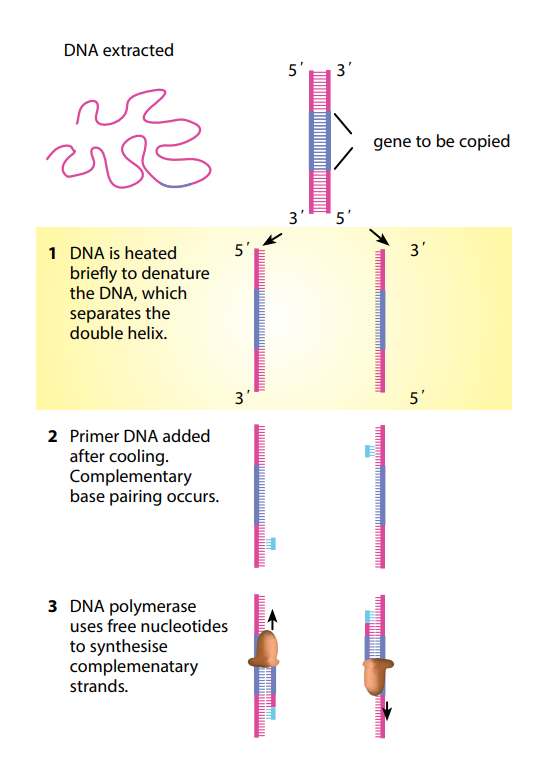
First, the DNA is denatured, usually by heating it. Th is separates the DNA molecule into its two strands, leaving bases exposed. Th e enzyme DNA polymerase is then used to build new strands of DNA against the exposed ones. However, DNA polymerase cannot just begin doing this with no ‘guidance’. A primer is used to begin the process. Th is is a short length of DNA, oft en about 20 base pairs long, that has a base sequence complementary to the start of the part of the DNA strand that is to be copied. Th e primer attaches to the start of the DNA strand, and then the DNA
polymerase continues to add nucleotides all along the rest of the DNA strand.
Once the DNA has been copied, the mixture is heated again, which once more separates the two strands in each DNA molecule, leaving them available for copying again. Once more, the primers fix themselves to the start of each strand of unpaired nucleotides, and DNA polymerase makes complementary copies of them.
The three stages in each round of copying need different temperatures.
- Denaturing the double-stranded DNA molecules to make single-stranded ones requires a high temperature, around 95 °C.
- Attaching the primers to the ends of the singlestranded DNA molecules (known as annealing) requires a temperature of about 65 °C.
- Building up complete new DNA strands using DNA polymerase (known as elongation) requires a temperature of around 72 °C. Th e DNA polymerases used for this process come from microorganisms that have evolved to live in hot environments.
Most laboratories that work with DNA have a machine that automatically changes the temperature of the mixture. The DNA sample is placed into a tube together with the primers, free nucleotides, a buff er solution and the DNA polymerase. Th e PCR machine is switched on and left to work. Th e tubes are very small (they hold about 0.05 cm3) and have very thin walls, so when the temperature in the machine changes, the temperature inside the tubes changes very quickly.
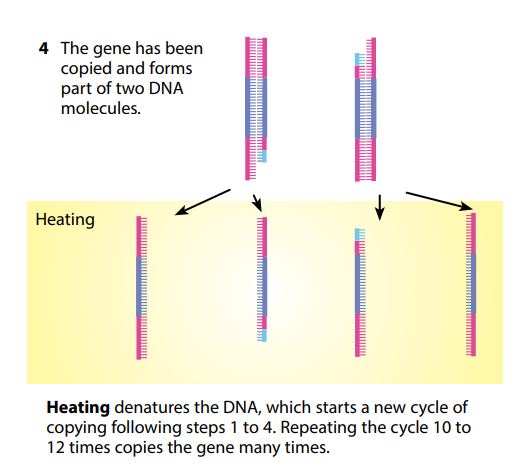
You can see that theoretically this could go on forever, making more and more copies of what might originally have been just a tiny number of DNA molecules. A single DNA molecule can be used to produce literally billions of copies of itself in just a few hours. PCR has made it possible to get enough DNA from a tiny sample – for example, a microscopic portion of a drop of blood left at a
crime scene.
How does PCR work?
The principles behind every PCR, whatever the sample of DNA, are the same.
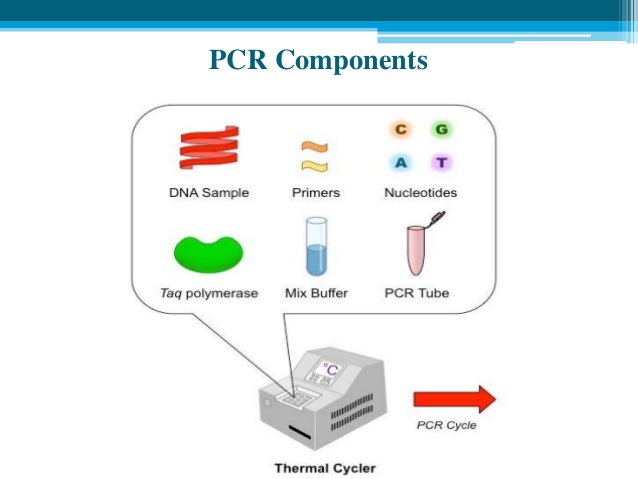
- Five core ‘ingredients’ are required to set up a PCR. We will explain exactly what each of these do as we go along.These are:
- the DNA template to be copied
- primers, short stretches of DNA that initiate the PCR reaction, designed to bind to either side of the section of DNA you want to copy
- DNA nucleotide bases? (also known as dNTPs). DNA bases (A, C, G and T) are the building blocks of DNA and are needed to construct the new strand of DNA
- Taq polymerase enzyme? to add in the new DNA bases
- buffer to ensure the right conditions for the reaction.
- PCR involves a process of heating and cooling called thermal cycling which is carried out by machine.
- Denaturing – when the double-stranded template DNA is heated with 96 °c temperature to separate it into two single strands.
- Annealing – when the temperature is lowered to 55 – 65°c to enable the DNA primers to attach to the template DNA.
- Extending – when the temperature is raised to 72 °c and the new strand of DNA is made by the Taq polymerase enzyme.
These three stages are repeated 20-40 times, doubling the number of DNA copies each time. A complete PCR reaction can be performed in a few hours, or even less than an hour with certain high-speed machines. After PCR has been completed, a method called electrophoresis can be used to check the quantity and size of the DNA fragments produced.
DNA Bands

What happens at each stage of PCR
1.Denaturing stage
- During this stage the cocktail containing the template DNA and all the other core ingredients is heated to 94-95⁰C.
- The high temperature causes the hydrogen bonds? between the bases in two strands of template DNA to break and the two strands to separate.
- This results in two single strands of DNA, which will act as templates for the production of the new strands of DNA.
- It is important that the temperature is maintained at this stage for long enough to ensure that the DNA strands have separated completely.
- This usually takes between 15-30 second
2.Annealing stage
- During this stage the reaction is cooled to 50-65⁰C. This enables the primers to attach to a specific location on the single-stranded template DNA by way of hydrogen bonding (the exact temperature depends on the melting temperature of the primers you are using).
- Primers are single strands of DNA or RNA? sequence that are around 20 to 30 bases in length.
- The primers are designed to be complementary? in sequence to short sections of DNA on each end of the sequence to be copied.
- Primers serve as the starting point for DNA synthesis. The polymerase enzyme can only add DNA bases to a double strand of DNA. Only once the primer has bound can the polymerase enzyme attach and start making the new complementary strand of DNA from the loose DNA bases.
- The two separated strands of DNA are complementary and run in opposite directions (from one end – the 5’ end – to the other – the 3’ end); as a result, there are two primers – a forward primer and a reverse primer.
- This step usually takes about 10-30 seconds.
3.Extending stage
- During this final step, the heat is increased to 72⁰C to enable the new DNA to be made by a special Taq DNA polymerase enzyme which adds DNA bases.
- Taq DNA polymerase is an enzyme taken from the heat-loving bacteria? Thermus aquaticus. 72⁰C is the optimum temperature for theTaq polymerase to build the complementary strand. It attaches to the primer and then adds DNA bases to the single strand one-by-one in the 5’ to 3’ direction.
- This bacteria normally lives in hot springs so can tolerate temperatures above 80⁰C.
- The bacteria’s DNA polymerase is very stable at high temperatures, which means it can withstand the temperatures needed to break the strands of DNA apart in the denaturing stage of PCR.
- DNA polymerase from most other organisms would not be able to withstand these high temperatures, for example, human polymerase works ideally at 37˚C (body temperature).
- The result is a brand new strand of DNA and a double-stranded molecule of DNA.
- The duration of this step depends on the length of DNA sequence being amplified but usually takes around one minute to copy 1,000 DNA bases (1Kb).
- These three processes of thermal cycling are repeated 20-40 times to produce lots of copies of the DNA sequence of interest.
- The new fragments of DNA that are made during PCR also serve as templates to which the DNA polymerase enzyme can attach and start making DNA.
- The result is a huge number of copies of the specific DNA segment produced in a relatively short period of time.
Gel Electrophoresis
Gel electrophoresis is a technique that is used to separate different molecules. It is used extensively in the analysis of proteins and DNA. This technique involves placing a mixture of molecules into wells cut into agarose gel and applying an electric field. The movement of charged molecules within the gel in response to the electric field depends on a number of factors. The most important are:
- Net (overall) charge – negatively charged molecules move towards the anode (+) and positively charged molecules move towards the cathode (–); highly charged molecules move faster than those with less overall charge.
- Size – smaller molecules move through the gel faster than larger molecules.
- Composition of the gel – common gels are polyacrylamide for proteins and agarose for DNA; the size of the ‘pores’ within the gel determines the speed with which proteins and fragments of DNA move.
Using Gel Electrophoresis to visualize the results of PCR
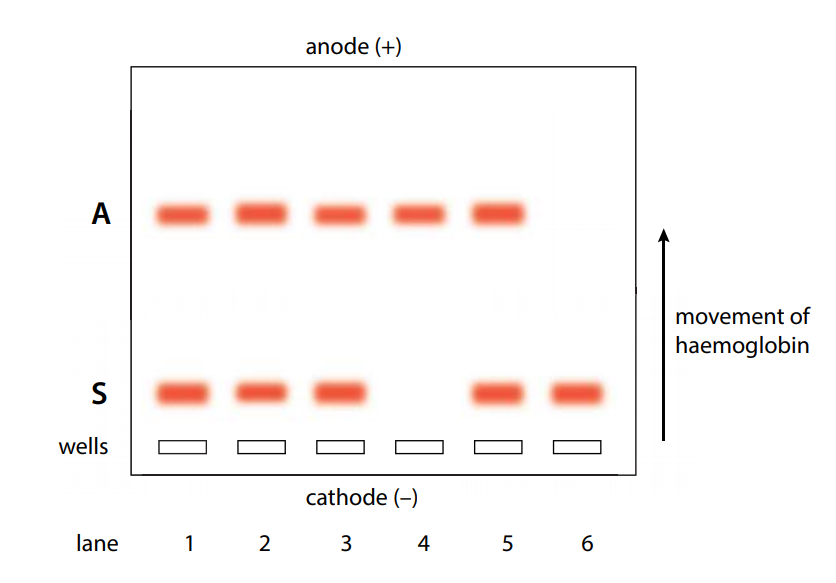
- The results of a PCR reaction are usually visualized (made visible) using gel electrophoresis. Gel electrophoresis is a technique in which fragments of DNA are pulled through a gel matrix by an electric current, and it separates DNA fragments according to size. A standard, or DNA ladder, is typically included so that the size of the fragments in the PCR sample can be determined.
- DNA fragments of the same length form a “band” on the gel, which can be seen by eye if the gel is stained with a DNA-binding dye. For example, a PCR reaction producing a 400400400 base pair (bp) fragment would look like this ona gel:
- Left lane: DNA ladder with 100, 200, 300, 400, 500 bp bands.
- Right lane: result of PCR reaction, a band at 400 bp.
- A DNA band contains many, many copies of the target DNA region, not just one or a few copies. Because DNA is microscopic, lots of copies of it must be present before we can see it by eye. This is a big part of why PCR is an important tool: it produces enough copies of a DNA sequence that we can see or manipulate that region of DNA.
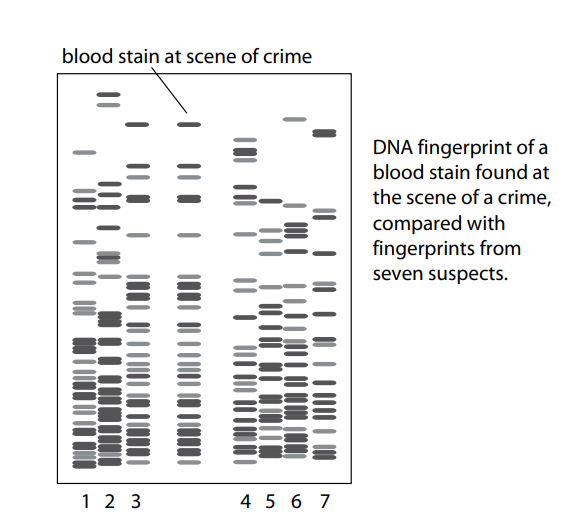
PCR is now routinely used in forensic science to amplify DNA from the smallest tissue samples left at the scene of a crime. Many crimes have been solved with the help of PCR together with analysis of DNA using gel electrophoresis.
Applications of PCR
- Applications in forensics
- In Parental testing
- PCR applied to diagnosis
- Detection of genetic diseases
- Detection of infectious diseases eg. Bacteria and Virus, Malaria
- PCR applied to identification of species
- For food authenticity : Meats, Seafood, Dairy products and milk, Cereals, Coffee, tea, chocolate, wine, fruit juices, Oils, Microbiota of food products
- Medicinal parts authenticity
- Acellular Cloning
- Reverse transcriptase PCR (RT-PCR)
- Quantitative PCR in real time (quantitative real-time PCR)
- Semi-quantitative or competitive PCR
So, in the end and in short, the polymerase chain reaction (PCR) is a method of making very large numbers of copies of DNA from very small quantities (even one molecule). In PCR, DNA is denatured by heat to separate the strands; a short length of DNA known as a primer attaches to one end of each strand so that DNA polymerase can start synthesising a complementary strand using free nucleotides (as in replication). The double-stranded copies of DNA are separated again and the process repeated many times to ‘bulk up’ the DNA. Heat-stable DNA polymerases are used in PCR; the first was Taq polymerase that is found in a thermophilic bacterium, Thermus aquaticus. And in the field of Genetic Engineering the uses of PCR is beyond our imagination.
Reference
This Article is completely based on by the lecture of Dr. Mohammad Zashim Uddin, Professor, Department of Botany, University of Dhaka
Some info and pictures have been added by author
Reference used for info: Cambridge International Biology Cource Book by Mary Jones
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka




