Author: Tahmina Mojumder
This article discusses
Recombinant DNA and gene cloning, plasmid and phase vectors, restriction enzymes, restriction maps and their properties, recombinant selection.
Recombinant DNA
Recombinant DNA is the common name for a segment of DNA that has been created by combining at least two fragments from two different sources.
Recombinant DNA is compacted into a host organism to produce new genetic combinations that are of value to science,prescript or medicine, agriculture, and industry.
Since, the center of all gene is genetics, the original goal of laboratory geneticists is to isolate, individualize, and manipulate genes. Although it is relatively easy to isolate a sample of DNA from a compilation of cells, it is finding a specific gene within this DNA sample can be compared to finding a needle in a haystack.
Each human cell contain about 2 metres of DNA. However, recombinant DNA technology can isolate a gene or any other segment of DNA to determine the nucleotide sequence. It has enabled to determine its nucleotide sequence, study its transcripts, and restore the modified sequence into living organisms.
The idea of recombinant DNA was first mentioned by Peter Lobban. He is the student of Stanford University medical School . In 1980,Paul Berg awarded the Nobel Prize in Chemistry for his work on nucleic acids with particular regard to recombinant DNA.
Recombinant DNA technology is based primarily on two other technologies,
Best safe and secure cloud storage with password protection
Get Envato Elements, Prime Video, Hotstar and Netflix For Free
Best Money Earning Website 100$ Day
#1 Top ranking article submission website
- DNA cloning and
- DNA sequencing.
i. Cloning is commenced in order to obtain the clone of one particular gene or DNA sequence of interest. The next step after cloning is to find and isolate that clone among other members of the library.once a segment of DNA has been cloned,its nucleotide sequence can be determind.Cloning is the method of propagating genetically identical copy of a cell or an organism.
There are three kind of artificial cloning. These are gene cloning, reproductive cloning and therapeutic cloning.
ii. DNA sequencing is the method of ascertaining the nucleic acid sequences the order of nucleotides in DNA and includes any method or technology . It is used to determine four bases (adenine, guanine, cytosine thymine). The first DNA sequences were obtained by academic researchers using laborious methods, in the early 1970 . Following the development of florescence based sequencing methods, including DNA sequencing. DNA sequencing has become easier and instantaneous commands faster.
Recombinant DNA technology
Recombinant DNA technology is used to create synthetic DNA by combining different genetic material (DNA) from different sources.
The recombinant DNA technology was first discovered in 1968 by Swiss microbiologist Werner Arber. It is inserting the desired gene into the genome of the host and involves the selection of the desired gene for administration within the host . Recombinant DNA technology follows a selection of the perfect vectors with which the gene must be integrated and recombinant DNA formed.
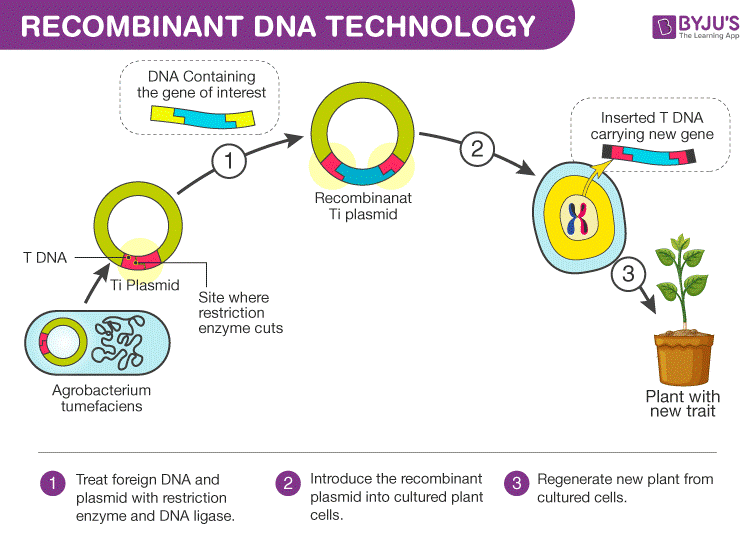
Process of Recombinant DNA Technology
The whole process of recombinant DNA technology involves multiple steps which are maintained in a certain sequence to generate the desired product.
Step-1. Isolation of Genetic Material.
The initial and first step in Recombinant DNA technology is to isolate the desired DNA in its purest form that is ,to free it from other macromolecules.
Step-2.Cutting the gene at the recognition sites.
The restriction enzymes play a major role in determining whether the desired gene has been inserted into the vector genome. These reactions are called ‘limiting enzyme digestions’.
Step-3. Amplify the gene copies through Polymerase chain reaction.
It is the process to translating DNA from a single copy into thousands to millions of copies once the proper gene of interest has been cut off using the restriction enzymes.
Step-4. Ligation of DNA Molecules.
In this step of Ligation, join of the two pieces –a cut piece of DNA and the vector together with the help of the enzyme DNA ligase.
Step-5. Insertion of Recombinant DNA into Host.
In this step, the recombinant DNA is introduced as a recipient host cell. This process is called Transformation. Once recombinant DNA enters into the host cell, it multiplies and is expressed in the form of proteins produced under favorable conditions.
Gene Cloning
Gene cloning is the process of separating a DNA sequence of interest for the purpose of making multiple copies of it and as a result, it is in which a gene of interest is located and copied (cloned) out of all the DNA obtained from an organism.
Gene cloning steps/Gene cloning process
The fundamental 7 steps involved in gene cloning are:
- DNA dissociation [gene of interest] need to be cloned.
- Insertion of isolated DNA into a suitable vectors for recombinant DNA formation.
- Recombinant DNA identification in a suitable organism known as host.
- Selection of converted host cells and identification of the clone containing the gene of interest.
- The quality and Expression of the gene introduced in the host.
- Isolation of multiple gene replication/Protein expressed by the genes.
- Isolated gene copy/protein purifier
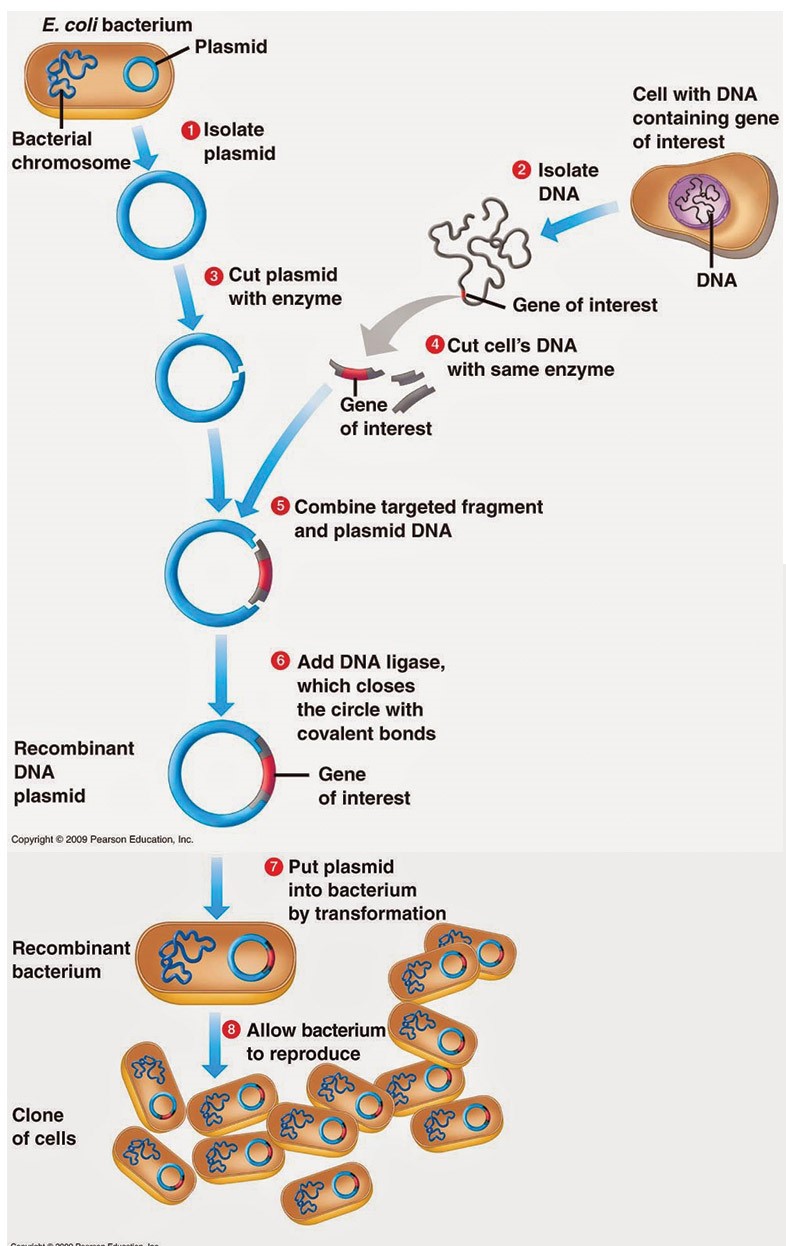
Plasmid and Phase vectors
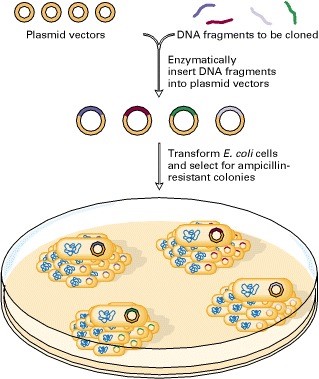
Basically, Plasmid is a small, round, double-stranded DNA molecule that is distinct from a cell’s chromosomal DNA and generally circular DNA sequences that are capable of replicating using the host cell’s replication equipment. Plasmids exist naturally in bacterial cells. Plasmid also occur in some eukaryotes and are especially important in treatment because they often carry genes for pathogenicity and drug-resistance. Plasmids can be transmitted to bacteria in the lab in a process called transformation. Plasmids used experimentally for these purposes are called vectors. Researchers who can insert a piece of DNA or a gene into a plasmid vector create a so-called recombinant plasmid. Plasmid vectors have a source of an replication that allows for semi-individual replication of the host’s plasmid.

Plasmid vectors are the earliest achievements of recombinant DNA technology, the figures of cloning and synthesis in E.coli of insulin and other hormones, interferon and animal virus antigens.
The term plasmid was coined in 1952 by the American molecular biologist Joshua Lederberg to refer to “any extrachromosomal hereditary determinant.
plasmids are determined of three major components that include
- Origin of replication (replicon)
- Polylinker (multiple cloning sites)
- Antibiotic resistance gene
Some of the other components of plasmids include
A promoter region – This is the component of plasmids that are involved in recruitment of transcriptional equipment.
Primer binding site – This is an abbreviated sequence of DNA on a single strand typically used for the purposes of PCR amplification or DNA sequencing.
Although plasmids share various general characteristics, different types in existence.
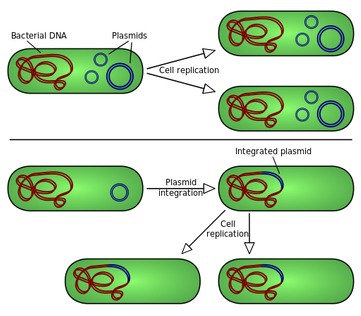
Molecular cloning and recombination tests are therefore repetitive processes which include :
- A DNA fragment is cloned into a plasmid cut with a consistency restriction enzyme
- Recombinant plasmids are converted into bacteria
- Bacteria are usually allowed to multiply liquid cultures
- Plenty of recombinant plasmid DNA is isolated from the bacterial culture
- Further manipulations (such as site directed mutagenesis or the introduction of another piece of DNA) are performed on the retrofit plasmid
- The modified plasmid is again converted to bacteria, before further manipulations or expression
Phage vector
Phage vectors consist of an essentially complete phage genome, often M13 phage, into which is inserted DNA encoding the protein or peptide of interest . Typically, the remainder of the phage genome is left unchanged and provides the other gene products needed for the phage life cycle These are typically plasmids with plasmid characteristics: round, double-stranded DNA with a replicated source, a selection marker, and a cloning site,where there is general sequence for the λ cos site.
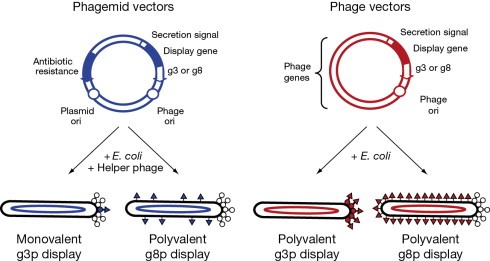
Bacteriophage
- Virus that infect bacteria is known as bacteriophage
- It was discovered by Frederick W. Twort in Great Britian and Felix d Herelle
- D Herelle coined the term bacteriophage meaning bacterial eater to describe the agents bacteriocidal activity
Why bacteriophage as a vector?
- It can accept very large fragment of foreign DNA
- Genetic Engineers have constracted numerous derivativesof phage vectors that have onlyone or two more sites for different restriction enzymes
- A stuffer piece in the phage is called substitution vectors because they are designed to have a slice removed.
- For example, Lambda phage , M13 phage, etc
Restriction enzyme, restriction maps and their properties
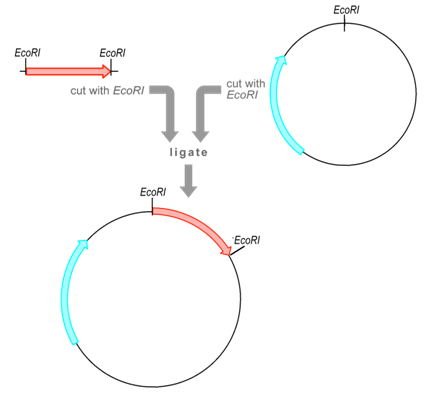
Restriction enzyme is an enzyme that pierces DNA into fragments at or near specific recognition sites within molecules.it is known as restriction sites.
Restriction enzyme originated from the studies of phage λ, which is a virus that infects bacteria, and the phenomenon of host-controlled restriction and modification of such bacterial phage . The phenomenon was first recognized in work done in the laboratories of Salvador Luria, Weigle and Giuseppe Bertani in the early 1950s. It was found that, for a bacteriophage λ which can grow well at one end of Escherichia coli, for example E. coli . Restriction enzymes apparently produced from a common ancestor and became widespread via horizontal gene transfer.
For Example ..
| Enzyme | Source | Recognition Sequence | Cut |
| EcoRI | Escherichia coli | 5’GAATTC
3’CTTAAG |
5′—G AATTC—3′
3′—CTTAA G—5′ |
| EcoRII | Escherichia coli | 5’CCWGG
3’GGWCC |
5′— CCWGG—3′
3′—GGWCC —5′ |
| BamHI | Bacillus amyloliquefaciens | 5’GGATCC
3’CCTAGG |
5′—G GATCC—3′
3′—CCTAG G—5′ |
| HindIII | Haemophilus influenzae | 5’AAGCTT
3’TTCGAA |
5′—A AGCTT—3′
3′—TTCGA A—5′ |
Properties of restriction enzyme
- Specificity
- Cleaves ds DNA only
- Can generate stickcy or blunt end.
- Palindromic sequence
- Size of recognition sequence
- Nomenclature of E
- Isoschizomer
- Neoschizomer
- Isocaudomer
Restriction map
A restriction map is a map that is known restriction sites within a sequence of DNA and is also a method used to map an unknown segment of DNA by breaking it into pieces and then identifying the locations of the breakpoints. Restriction mapping requires the use of restriction enzymes. Restriction maps are used as a reference to comparatively other shorts parts of engineered plasmids or DNA, and sometimes for longer genomic DNA. There are other ways of mapping features on DNA for longer length DNA molecules, such as mapping by transduction.Map can be constructed.
Recombinant selections
Selection is the process in which cells that have not taken up DNA are selectively killed, and only those cells that can actively replicate DNA.It contains the selectable marker gene encoded by the vector are able to survive.It is essential to identify those cells which received rDNA molecule , after the introduction of recombinant DNA into the host cells.
Artificial selection is used in the field of microbial genetics and especially molecular cloning. Molecular cloning is a set of epimerical methods in molecular biology that are used to congregate recombinant DNA molecules . It is used to direct their replication within host organisms.
Reference
Restriction enzyme from Wikipedia
Revised by
Abulais Shomrat on 12 February, 2021.
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka






I do not even know how I ended up here, but I thought this post was great. I don’t know who you are but definitely you’re going to a famous blogger if you aren’t already 😉 Cheers!
I loved as much as you will receive carried out right here. The sketch is tasteful, your authored subject matter stylish. nonetheless, you command get got an edginess over that you wish be delivering the following. unwell unquestionably come further formerly again as exactly the same nearly very often inside case you shield this hike.
Thank you so much for your Valuable comment. We wish to hear from you again.
I was recommended this website by my cousin. I am not sure whether this post is written by him as nobody else know such detailed about my trouble. You are amazing! Thanks!
Thank you so much for your Valuable comment. We wish to hear from you again.
Nice blog here! Also your site loads up fast! What host are you using? Can I get your affiliate link to your host? I wish my web site loaded up as quickly as yours lol
Simply wish to say your article is as amazing. The clearness in your post is just nice and i could assume you’re an expert on this subject. Well with your permission let me to grab your feed to keep updated with forthcoming post. Thanks a million and please carry on the gratifying work.
Thank you so much for your Valuable comment. We wish to hear from you again.
Somebody essentially lend a hand to make significantly posts I might state. That is the very first time I frequented your web page and up to now? I surprised with the research you made to create this particular put up amazing. Excellent job!