The first step in sequencing a genome is to divide the individual chromosomes (in eukaryotes) or whole DNA (in prokaryotes) in an ordered manner into smaller and smaller pieces that ultimately can be sequenced. That is, one begins by creating a genomic library of fragmented DNA parts.
However, at some stage, the different clones (after cloning of the fragmented parts) have to be ordered into a physical map corresponding to that found in the intact organismal genome. The magnitude of this task depends on the average size of the cloned fragment: the larger the fragment, the fewer clones that have to be ordered.
Restriction Enzymes: The Biological Knives
The ability to clone, cut and sequence essentially any gene or other DNA sequence of interest from any species depends on a special class of enzymes called restriction endonucleases (from the Greek term éndon meaning “within”; endonucleases make internal cuts in DNA molecules).
Many endonucleases make random cuts in DNA, but the restriction endonucleases are site-specific, and Type II restriction enzymes are the most used biological knives in the laboratory to cut or cleave DNA.
Type II restriction enzymes cleave DNA molecules only at specific nucleotide sequences called restriction sites. Type II restriction enzymes cleave DNA at these sites regardless of the source of the DNA.
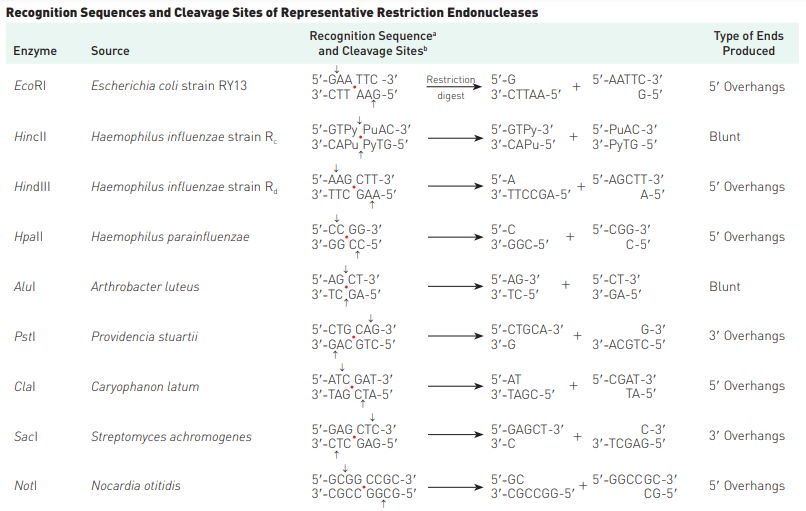
- An interesting feature of restriction endonucleases is that they commonly recognize DNA sequences that are palindromes—that is, nucleotide-pair sequences that read the same forward or backward from a central axis of symmetry. Simply can be put as, the sequences are palindromic i.e. from 5′ to 3′ (or, 3′ to 5′) direction, they are same. For example, in case of EcoRI, both strands in 5′ to 3′ stand as GAATTC.
- In addition, a useful feature of many restriction nucleases is that they make staggered cuts; that is, they cleave the two strands of a double helix at different points (Figure 1). (Other restriction endonucleases cut both strands at the same place and produce bluntended fragments.)
How restriction enzymes are named?
The restriction endonucleases are named by using the first letter of the genus and the first two letters of the species that produces the enzyme. If an enzyme is produced only by a specific strain, a letter designating the strain is appended to the name. The first restriction enzyme identified from a bacterial strain is designated I, the second II, and so on.
Thus, restriction endonuclease EcoRI is produced by Escherichia coli strain RY13. Hundreds of restriction enzymes have been characterized and purified; thus, restriction endonucleases that cleave DNA molecules at many different DNA sequences are available. For an extensive list of restriction enzymes, start here: List of Restriction Enzyme Cutting Site: A.
Discovery & purposes of restriction enzymes
Restriction endonucleases were discovered in 1970 by Hamilton Smith and Daniel Nathans (see A Milestone in Genetics: Restriction Endonucleases on the Student Companion site). They shared the 1986 Nobel Prize in Physiology or Medicine with Werner Arber, who carried out pioneering research that led to the discovery of restriction enzymes. The biological function of restriction endonucleases is to protect the genetic material of bacteria from “invasion” by foreign DNAs, such as DNA molecules from another species or viral DNAs. As a result, restriction endonucleases are sometimes referred to as the immune systems of prokaryotes.
How a bacterium protects its DNAs restriction sites from being cut by its own restriction enzyme?
All cleavage/restriction sites in the DNA of an organism must be protected from cleavage/cutting by the organism’s own restriction endonucleases; otherwise the organism would commit suicide by degrading its own DNA.
In many cases, this protection of endogenous cleavage sites is accomplished by methylation of one or more nucleotides in each nucleotide sequence that is recognized by the organism’s own restriction endonuclease. Methylation occurs rapidly after replication, catalyzed by site-specific methylases produced by the organism.
Each restriction endonuclease will cleave a foreign DNA molecule into a fixed number of fragments, the number depending on the number of restriction sites in the particular DNA molecule.
The Availability of Restriction Sites
Type II restriction endonucleases have target sites which are 4–8 bp (=base pairs) in length. If all bases i.e. A, T, C, G are equally frequent in a genome or DNA molecule, we would expect a tetranucleotide (a restriction site of 4 bp) to occur and cut the sequence on average every 44 (i.e. 256) nucleotide pairs in a long random DNA sequence. Similarly,
- a hexanucleotide (a restriction site of 6 bp) would occur and make fragments every 46 (i.e. 4096) bp and
- an octanucleotide (a restriction site of 8 bp) every 48 (i.e. 65 536) bp.
Remember, if restriction site is found after every ‘n’ number of base pairs, the length of the fragments would also be ‘n’ base pairs.
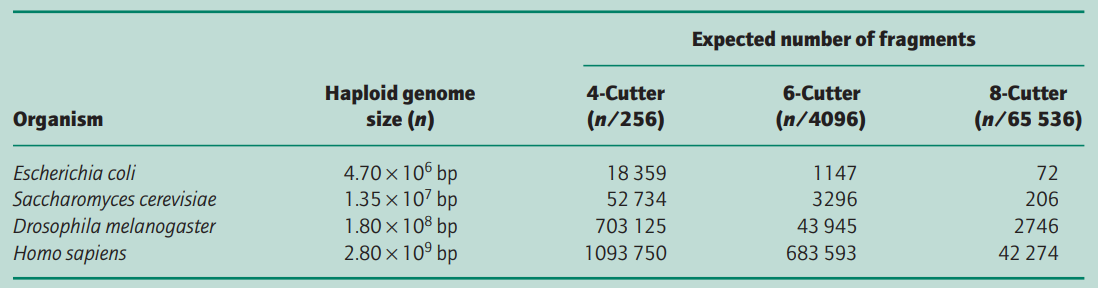
Table 1 shows the expected number of fragments that would be produced when different genomes are digested completely with different restriction endonucleases.
In practice, the actual number of fragments/restriction sites is quite different because the distribution of nucleotides is non-random and most organisms do not have an equal number of the four bases; e.g. human DNA has overall only 40% G + C, 60% A + T content and the frequency of the dinucleotide CpG is only 20% of that expected.
In addition, many cytosine residues are methylated and this can prevent restriction endonuclease digestion. Thus the enzyme NotI, for which the restriction site is GCGGCCGC (8 bp), cuts human DNA into fragments of average size 1000– 1500 kb rather than the 65 kb (28 = 65 536 bp) expected. Similarly, the Escherichia coli genome is cut by NotI into only 20 fragments, not the 72 expected from Table 1 (Smith et al. 1987).
Good to know
- CpG means a site where cytosine (C) lies next to guanine (G) in the DNA sequence. (The p indicates that C and G are connected by a phosphodiester bond.) Methylation of DNA occurs at any CpG site.
- Methylation means addition of methyl group to nucleotide. If this remains attached to a nucleotide, it becomes uncuttable with restriction enzyme (irrespective of the presence of restriction sites).
- Restriction digestion means cutting sequences with Restriction enzymes.
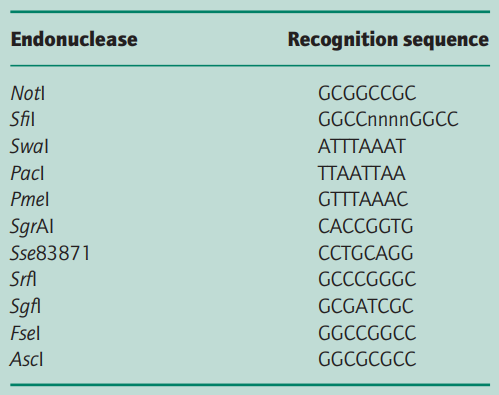
Gelfand and Koonin (1997) have analysed those bacterial genomes that have been completely sequenced and have found that short palindromic sequences, like restriction endonuclease recognition sites, are low in number at a statistically significant level.
The average size of DNA fragment produced by digestion with restriction enzymes with 4- and 6-base recognition sequences is too small to be of much use for preparing gene libraries except in special circumstances. Even enzymes with 8-base recognition sequences may not be of particular value because, although the average size of fragment should be 65.5 kb, in the case of S. pombe, in practice the fragments range in size from 4.5 kb to 3.5 Mb (Fan et al. 1989).
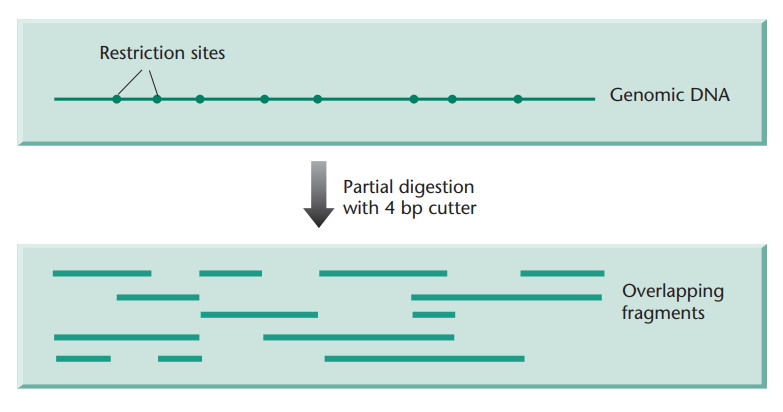
If more uniform-sized fragments are required, it is usual to partially digest the target DNA with an enzyme with a 4-base recognition sequence. The partial digest then can be fractionated to separate out fragments of the desired size. Because the DNA is randomly fragmented there will be no exclusion of any sequence. Furthermore, clones will overlap one another (Fig. 2) and this is particularly important when trying to order different clones into a map.
Some introns encode endonucleases that are site specific and have 18–30 bp recognition sequences (Dujon et al. 1989). These endonucleases can be used to produce a very limited number of fragments, some or all of which are produced by cleavage within related genes.
References
- Fan J-B., Chikashige Y., Smith C.L., Niwa O., Yanagida M. & Cantor C.R. (1989) Construction of a Not I restriction map of the fission yeast Schizosaccharomyces pombe genome. Nucleic Acids Research 17, 2801–2818.
- Gelfand M.S. & Koonin E.V. (1997) Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Research 25, 2430–2439.
- Principles of Genetics (Sixth Edition) by Snustad & Simmons.
- Principles of Genome Analysis & Genomics (Third Edition) by Primrose & Twyman.
- Smith C.L., Econome J.G., Schutt A., Klco S. & Cantor C.R. (1987) A physical map of the Escherichia coli K12 genome. Science 236,1448–1453.
 Plantlet The Blogging Platform of Department of Botany, University of Dhaka
Plantlet The Blogging Platform of Department of Botany, University of Dhaka




